 |
 |
| Anim Biosci > Volume 32(1); 2019 > Article |
|
Abstract
Objective
Recent studies have demonstrated that lin-28 homolog B (LIN28B)/miRNA let-7 (let-7) plays a role in the regulation of pubertal onset in mammals. However, the role of LIN28B/let-7 in the onset of ovine puberty remains unknown. We cloned the Duolang sheep Lin28B cDNA sequence, detected the expression change of LIN28B, let-7a and let-7g in hypothalamus, pituitary and ovary tissues at three different pubertal stages.
Methods
The reverse transcriptase polymerase chain reaction (RT-PCR) was used to clone the cDNA sequence of LIN28B gene from Duolang sheep and the bioinformatics methods were applied to analyze the amino acid sequence of LIN28B protein. The mRNA expression levels of the LIN28B gene at different pubertal stages were examined by real time RT-PCR.
Results
LIN28B cDNA of Duolang sheep was cloned, and two transcripts were obtained. The amino acid sequence of transcript 1 shares 99.60%, 98.78%, and 94.80% identity with those of goat, wild yak and pig, respectively. Strong LIN28B mRNA expression was detected in the hypothalamus, pituitary, ovary, oviduct and uterus, while moderate expression was found in the liver, kidney, spleen and heart, weak expression was observed in the heart. No expression was found in the lungs. Quantitative real-time PCR (QPCR) and western-blot analysis revealed that the LIN28B was highly expressed in the hypothalamus and ovary at prepuberty stages, and this expression significantly decreased from the prepuberty to puberty stages (p<0.05). Markedly increased levels of mRNA expression were detected in the pituitary from prepuberty to puberty (p<0.05) and then significantly decreased from puberty to postpuberty (p<0.05). The expression levels of let-7a and let-7g showed no significant changes among different pubertal stages (p>0.05).
Puberty is a key stage in which animals attain fertility for the first time and involves a complex series of events that are governed by the activation of the hypothalamic-pituitary-gonadal axis. The age of puberty is a moderately heritable trait, with an average heritability of 0.32 [1,2]; normal or disturbed pubertal development is mainly determined by genetic factors [3,4]. The onset of puberty is closely related to changes in the transcription and expression levels of related genes, and lin-28 homolog B (LIN28B) is a particularly important gene associated with the onset of puberty [5ŌĆō7].
The LIN28 family includes two homologous members, LIN28A and LIN28B, each with a similar domain structure and function [8ŌĆō10]. LIN28B, which contains a cold shock domain (CSD) and a retroviral-type CCHC zinc finger motif, was first cloned and identified as an over-expressed factor in hepatocellular carcinoma cells [10]. LIN28B blocks miRNA let-7 (let-7) expression, whereas let-7 negatively regulates LIN28B expression by binding to the 3ŌĆ▓UTR of LIN28B, thereby establishing a double negative feedback loop. However, the expression patterns of LIN28B compared to those of let-7 are not completely reciprocal [11ŌĆō14].
In rodents, expression of LIN28B and let-7 in the hypothalamus-pituitary-gonad (HPG) tissues has been widely studied, the results show that LIN28B is expressed at a high level in the hypothalamus and testes, and that this expression is reversed in the ovary [15ŌĆō17]. In humans, LIN28B is widely expressed in adult normal tissues, and its expression level is highest in the testis and placenta [10]. In adult rats, the LIN28B mRNA expression level is higher in the testis, placenta and hypothalamus, whereas no expression is detected in the ovary [14].
Numerous studies have reported the vital correlation between LIN28B/let-7 and the onset of puberty in mice, monkeys, among other species, but there are few studies on the role of LIN28B/let-7 in the onset of ovine puberty [13].
Duolang sheep are a typical representative of an early maturing breed in Xinjiang, and its age of puberty is only 3ŌĆō4 months under the same ecological conditions [18]. In the present study, the LIN28B cDNA sequence from Duolang sheep was cloned and sequenced, and its expression profile was characterized in ten tissues. The expression levels of LIN28B, let-7a and let-7g were detected in the hypothalamus, pituitary and ovary at three different pubertal stages.
This work was conducted in accordance with the specifications of the Ethics Committee of Tarim University of Science and Technology.
Fifteen female Duolang sheep were housed, including five prepubertal, five pubertal, and five postpubertal sheep, all of which were obtained from the same line and reared under similar conditions on a farm in Xinjiang, China. We determined pubertal sheep in a study of female sheep by detecting estrus and changes in the appearance of the vulva [19,20]. Sheep were deeply anesthetized by intravenous administration of 3% pentobarbital sodium (30 mg/kg; Solarbio, P8410, Beijing, China) and sacrificed by exsanguination at a healthy physiological stage. The hypothalamus, pituitary, ovary, oviduct, uterus, liver, heart, kidney, lung and spleen were surgically removed and snap frozen in liquid nitrogen for mRNA extraction.
Total RNA was isolated from the hypothalamus, pituitary, ovary, oviduct, uterus, liver, heart, kidney, lung and spleen by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to a standard extraction protocol. The integrity of total RNA was evaluated on 1.5% agarose gels containing formaldehyde and ethidium bromide. First-strand cDNA was synthesized as follows: a mixture of 2 ╬╝g of total RNA, 1 ╬╝L of the oligo(dT)15 primer and 1 ╬╝L of dNTPs (2.5 mM each) was heated for 5 min at 65┬░C and chilled on ice; then, 4 ╬╝L of 5├Ś first-strand buffer, 0.5 ╬╝L of RNase Inhibitor (40 ╬╝L, TaKaRa, Dalian, China) and 1 ╬╝L of M-MLV enzyme (200 U/╬╝L, TaKaRa, China) were added, and the reaction was incubated for 50 min at 42┬░C, followed by 5 min at 95┬░C to inactivate the reverse transcriptase. The reaction was finally stored at ŌłÆ20┬░C for subsequent gene cloning and mRNA expression analysis.
LIN28B mRNA sequences of sheep and cattle (GenBank: JQ277700.1, XM_012182382.2 and XM_024997174.1, respectively) were used to design the primers for cloning (Table 1), and LIN28B cDNA was obtained from total RNA isolated from the ovary tissues of Duolang sheep. The polymerase chain reaction (PCR) was performed in a 25 ╬╝L total volume, containing 1 ╬╝L of first-strand cDNA template, 0.5 ╬╝L of each primer (10 ╬╝M), and 12.5 ╬╝L of PCR mix (TaKaRa, China). Reverse transcriptase-PCR (RT-PCR) with primers (Table 1) was performed at 95┬░C for 5 min, followed by 35 cycles of 95┬░C for 30 s, annealing for 30 s and 72┬░C for 1 min, followed by one cycle of 72┬░C for 5 min. The amplified PCR product was purified and ligated into pMD18-T (TaKaRa, China) and then transformed into competent Escherichia coli DH5╬▒ cells. At least four positive clones were sequenced with an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA).
The RT-PCR was performed to examine the LIN28B expression profile in ten tissues, including the hypothalamus, pituitary, ovary, oviduct, uterus, liver, heart, kidney, lung and spleen. PCR amplifications were performed in a 25 ╬╝L volume containing 1 ╬╝L of first-strand cDNA template, 0.5 ╬╝L of each primer (10 ╬╝M), and 12.5 ╬╝L of PCR mix (TaKaRa, China). For tissue expression analysis, beta-actin (ACTB) was used as the internal control and amplified with the specific primers ACTB-F and ACTB-R (Table 1). PCR products for ACTB and LIN28B were run separately on 1.5% agarose gels. The identity of each amplified fragment was confirmed by direct sequencing on an ABI 3730 sequencer (Applied Biosystems, USA).
The expression of LIN28B, let-7a, and let-7g in the hypothalamus, pituitary and ovary tissues of Duolang sheep at three different pubertal stages was assessed by quantitative real-time PCR (qPCR).
For LIN28B mRNA expression, ACTB was selected as the reference gene, and its quantification cycle was used to normalize the mRNA levels of the target gene. Primers were designed to span exonŌĆōexon boundaries to ensure specificity, which was further ensured according to dissociation curve, analysis and sequencing. The following primer sequences were used for LIN28B and ACTB: LIN28B F: 5ŌĆ▓-aggaagcgaaagaagaccca -3ŌĆ▓, R: 5ŌĆ▓-gcacttctttggctgaggag -3ŌĆ▓; and ACTB F: 5ŌĆ▓-ttccagccttccttcctg -3ŌĆ▓, R: 5ŌĆ▓-ccgtgttggcgtagaggt -3ŌĆ▓.
For let-7a and let-7g quantification, cDNA was synthesized by using a One-Step miRNA RT Kit. The mature sequences of let-7a and let-7g were obtained by using miRBase, and these sequences were used to design the primers. The forward primers for let-7a and let-7g were designed, while the reverse primers for let-7a and let-7g were included in the Sino Gene One-step miRNA RT Kit; U6 was selected as a reference gene. The following primer sequences were used for let-7a, let-7g and U6 snRNA: let-7a-F: 5ŌĆ▓-gctgaggtagtaggttgtatagtt -3ŌĆ▓; let-7g- F: 5ŌĆ▓-cgtgaggtagtagtttgtacatgt -3ŌĆ▓; and U6 - F: 5ŌĆ▓-ctcgcttcggca gcacatat -3, R: 5ŌĆ▓-aacgcttcacgaatttgcgt -3ŌĆ▓.
PCR was performed in a total volume of 20 ╬╝L, containing 1 ╬╝L of first-strand cDNA template, 7.5 ╬╝L of 2├Ś SG Green qPCR Mix, and 0.25 ╬╝L of each primer. PCR was performed under the following conditions: 95┬░C for 10 min, followed by 40 cycles at 95┬░C for 20 s and 60┬░C for 30 s, then at 95┬░C for 15 s, 60┬░C for 30 s, and 95┬░C for 15 s. Each sample was run three times on three separate assays. The expression levels of LIN28B, let-7a, and let-7g were calculated by the 2ŌłÆ╬ö╬öCT method [21].
Total protein extraction: the tissue was frozen in liquid nitrogen, and 100 mg of the tissue was added to 500 ╬╝L of RIPA lysate and mixed well, and then, the mixture was placed on ice for 30 min and homogenized; the mixture was then centrifuged at 12,000 rpm and 4┬░C for 15 min, and the supernatant was collected.
Protein concentrations were quantified with the Bicinchoninic Acid Protein Assay Kit (CW Biotech, Beijing, China), and the samples were incubated overnight with one of the following primary antibodies at 4┬░C: anti-Lin28B antibody (1:1,000, ab46020, Abcam, Boston, MA, USA) or anti-╬▓-actin antibody (1:1,000).
The secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit antibodies (1:5,000). An enhanced chemiluminescence kit (Millipore, Billerica, MA, USA) was used for detection.
For qPCR analysis, the mRNA levels of LIN28B, let-7a, and let-7b were expressed as the means┬▒standard error, and the means of the paired groups were analyzed by a paired DuncanŌĆÖs t-test. The differences were considered significant when p<0.05. All statistical analyses were performed by using the general linear model procedure of SAS (SAS version 9.2., Cary, NC, USA).
After RT-PCR with the primers B1ŌĆōB5, five fragments were sequenced and assembled according to the overlap sequences; thus, transcript 1 was obtained, which was 6,630 bp in size and contained 1,396 bp of the 5ŌĆ▓ UTR, 744 bp of the entire coding region and 4,490 bp of the 3ŌĆ▓ UTR. After RT-PCR with the primers B1ŌĆōB4 and B6, five fragments were sequenced and assembled according to the overlap sequences; thus, transcript 2 was obtained, which was 5,271 bp in size and contained 13 bp of the 5ŌĆ▓UTR, 768 bp of the entire coding region and 4,490 bp of the 3ŌĆ▓ UTR. The alignment of the two transcripts indicated that these sequences had different coding regions and 5ŌĆ▓ UTRs.
The amino acid sequences were deduced by using DNAman 6.0, the entire coding region of transcript 1 was predicted to encode an- 247 amino acid protein with a calculated molecular mass of 26.67 kDa, and the entire coding region of sheep transcript 2 was predicted to encode an- 255 amino acid protein with a calculated molecular mass of 27.55 kDa. The isoelectric points of LIN28B isoform 1 and isoform 2 were 8.78 and 9.08, respectively. The identities of the corresponding protein sequences were 96.08% between the two transcripts, and the transmembrane protein prediction by the TMHMM program showed that the LIN28B protein was not a transmembrane protein. The signal peptide prediction by Signalp showed that the LIN28B protein was not a signal peptide. Protein sequence analysis in the SMART database revealed that the proteins of the two transcripts contained N-terminal CSDs and a pair of C-terminal retroviral-type CCHC zinc finger domains, and the sequences of the CSDs and CCHC domains were conserved among mammalian species.
The LIN28B isoform1 sequence of Duolang sheep shares 99.60%, 98.78%, and 94.80% identity with those of goat (XP_ 005684673.1), wild yak (XP_005901662.1), and pig (XP_0209 35097.1), respectively (Figure 1). The LIN28B isoform1 sequence of Duolang sheep also shares 90.80%, 81.67%, 81.67%, and 76.92% with those of human (AAZ38897.1), Japanese quail (XP_015714194), chicken (AAZ38896.1), and house mouse (AAZ38894.1), respectively (Figure 2).
The phylogenetic relationship of LIN28B proteins among different species was investigated by using the Neighbor-Joining method, and the results clearly showed that the two LIN28B protein of Duolang sheep were clustered with those of goat, wild yak and pig; the house mouse and human LIN28B proteins formed a closely related group, while the Japanese quail and chicken LIN28B proteins formed another distinct group.
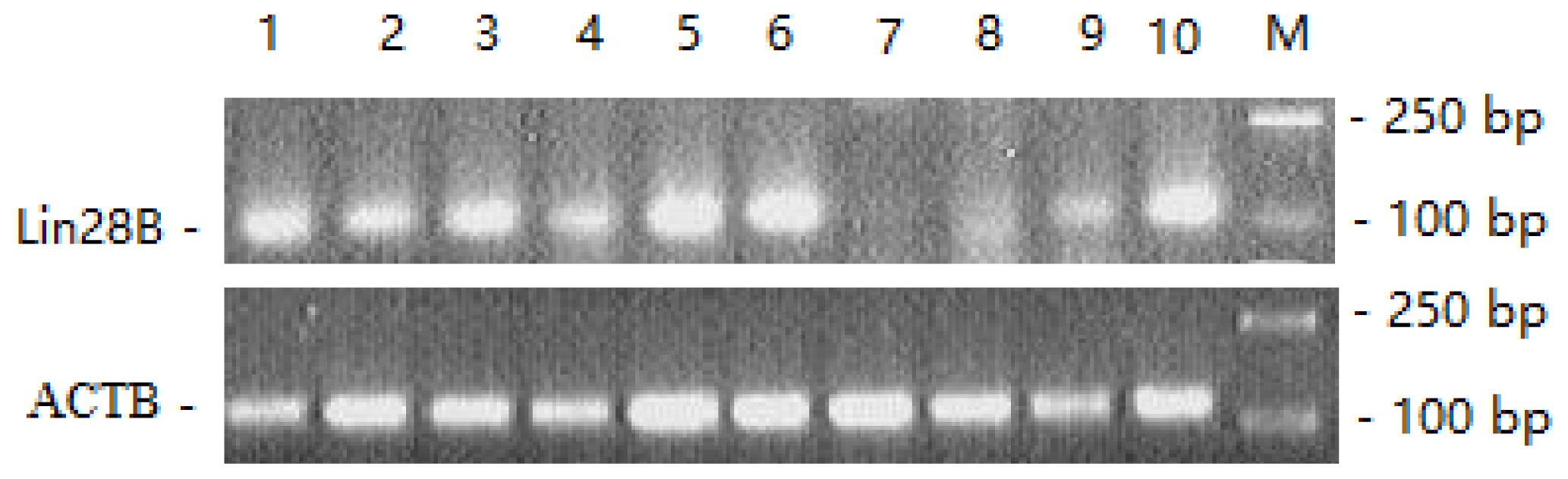
The expression profiles of LIN28B mRNA in ten tissues of Duolang sheep at the prepubertal stage were assessed (Figure 3). Strong expression of LIN28B mRNA was found in the hypothalamus, pituitary, ovary and oviduct; moderate expression was found in the spleen, kidney and liver; weak expression was found in the heart; and no expression was found in the lungs.
The expression patterns of LIN28B in the hypothalamus, pituitary and ovary at three different pubertal stages were detected using real-time PCR.
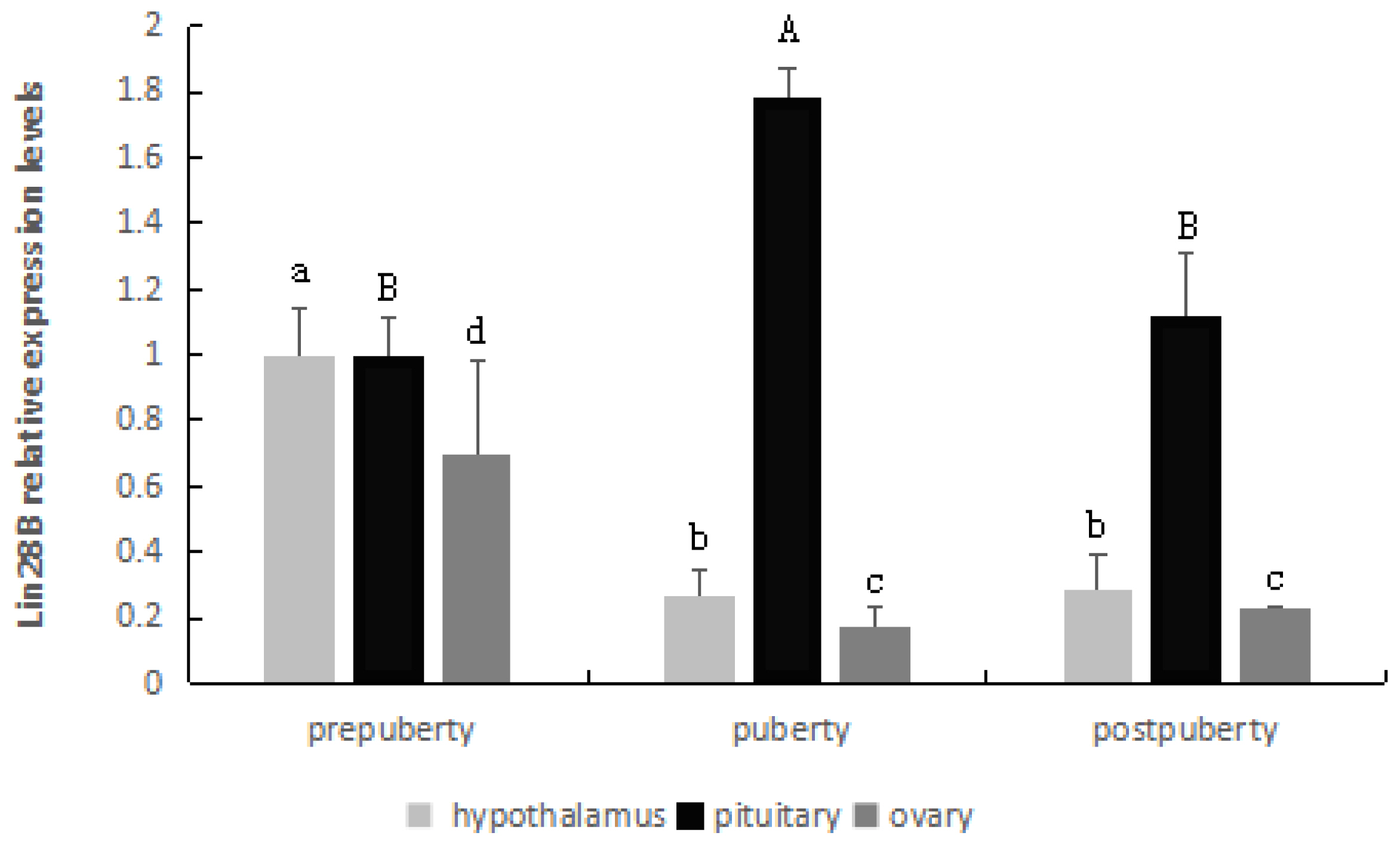
As observed in Figure 4, the LIN28B gene was highly expressed in the hypothalamus and ovary at prepuberty stages, and its expression was significantly decreased from prepuberty to puberty (p<0.05) but was extremely stable from puberty to postpuberty (p>0.05).
In the pituitary, there was a significantly increased level of mRNA expression from prepuberty to puberty (p<0.05), followed by a statistically significant decrease from puberty to postpuberty (p<0.05).
To investigate whether the expression of let-7a and let-7g changes across the pubertal transition, the expression levels of let-7a and let-7g were detected using qPCR. For let-7a, a trend toward increased expression in the hypothalamus and pituitary was observed from prepuberty to postpuberty but did not reach a significant level (p>0.05). The expression of let-7a in the ovary showed some changes among different pubertal stages, but there was no regularity and the levels did not reach significance (p> 0.05). Similarly, the expression of let-7g in the pituitary gradually increased from prepuberty to postpuberty but did not reach a significant level (p>0.05).
The protein expression pattern of LIN28B in the hypothalamus, pituitary and ovary at three different pubertal stages was detected using Western blotting (Figures 5, 6). As shown in Figure 5, the LIN28B protein was highly expressed in the hypothalamus and ovary, but significantly decreased from prepuberty to puberty (p<0.05). Expression of the Lin28B protein in the pituitary showed some changes among different pubertal stages, but the levels did not reach significance (p>0.05).
Alternative splicing of eukaryotic transcripts is a mechanism that enables cells to generate vast protein diversity from a limited number of genes [22]. In the present study, the LIN28B cDNA sequence of Duolang sheep was cloned and analyzed, and subsequently, we detected two LIN28B transcripts that differed in the 5ŌĆ▓ UTR and partial coding regions, the results of the present study are consistent with that of goat [23]. The 3ŌĆ▓ UTRs of the two transcripts were long and included let-7 microRNA complementary sites, similar to the human and goat sequences [10,23].
The LIN28B protein sequence was highly conserved among mammals [23], and in the present study, the LIN28B protein of Duolang sheep showed relatively high homology with those of other mammals. The amino acid sequence of Duolang sheep LIN28B transcript 1 shares 99.60%, 98.78%, and 94.80% identity with those of goat, wild yak and pig, respectively, and phylogenetic analysis showed that the phylogenetic tree conformed to evolutionary laws. Similar homology of LIN28B was found between goat, human and other known vertebrates [10,23].
The expression patterns of LIN28B were detected in several vertebrates [14,17]. LIN28B was expressed in many developing tissues and was involved in the biosynthesis of the let-7 family and the onset of mammalian puberty [24]. Expression of LIN28B was highest in the testis, placenta and hypothalamus of human and rats [10,23]. In goats, LIN28B was expressed in the hypothalamus, lung and spleen [25]. In the present study, we detected LIN28B mRNA expression in most tissues of Duolang sheep; strong LIN28B expression was found in the hypothalamus, pituitary, ovary and oviduct; moderate expression was found in the spleen, kidney and liver; weak expression was detected in the heart. The LIN28B expression differences in different tissues may exist in different species, and the LIN28B gene was mainly expressed in the hypothalamus, pituitary, ovary and testis [26]. From the above results, we concluded that LIN28B gene was distributed in the hypothalamus-pituitary-gonadal axis (HPGA), which participates in the onset of puberty regulation. The LIN28/let-7 system including LIN28A/LIN28B and let-7 miRNAs, has emerged as putative regulator of puberty [15,17]. In male and female rats, LIN28B displayed high hypothalamic expression during the neonatal period, which was markedly decreased during the infantile-to-juvenile transition and reached minimal levels before/around puberty. A similar puberty-related decline was observed for LIN28B in the hypothalamus of monkeys [14]. In the present study, LIN28B was highly expressed in the hypothalamus at prepuberty stages, but significantly decreased from prepuberty to puberty, and these results were consistent with those of a previous study [14].
In female mice, expression of LIN28B decreases in the ovary prior to the onset of puberty, and neither let-7a nor let-7g levels significantly changed [13]. In the present study, LIN28B expression was decreased in the ovaries prior to the onset of puberty, and neither let-7a nor let-7g levels significantly changed. Additionally, no statistically significant changes were observed in let-7a and let-7g levels in the hypothalamus and pituitary tissues among different pubertal stages. The present results showed that although LIN28B expression was decreased in the hypothalamus and ovary prior to the onset of puberty, neither the let-7a nor let-7g levels significantly increased. The data suggest that the sheep LIN28B effects within the HPG axis may be miRNA independent, which will be an important area for further study.
In summary, the results of the present study showed that LIN28B mRNA expression was detected in most of the tissues of Duolang sheep: strong expression of LIN28B was detected in the hypothalamus, pituitary, ovary and oviduct; moderate expression was found in the spleen, kidney and liver; weak expression was observed in the heart. LIN28B expression was distributed in the HPGA, and although LIN28B expression was decreased in the hypothalamus and ovary prior to the onset of puberty, neither let-7a nor let-7b levels were significantly increased. These data suggest that the sheep LIN28B effects within the HPG axis may be miRNA independent and that the LIN28B effects within the HPG axis will be an important area for further study.
ACKNOWLEDGMENTS
The present study was financially supported through grants from the National Natural Science Fund Project in China (31660652), and the Corps of Science and Technology Plan Projects (2015AG013), and Tarim Animal Husbandry Science and Technology Key Laboratory Open Topics (201504), and the Principal Fund Project of Tarim University (TDZKGG 201703).
Figure┬Ā1
Alignment of the amino acid sequences of the predicted Duolang sheep lin-28 homolog B protein with those of pig (XP_020935097.1), goat (XP_005684673), and wild yak (XP_005901662.1).

Figure┬Ā2
Alignment of the amino acid sequences of the predicted Duolang sheep lin-28 homolog B protein with those of human (AAZ38897.1), Japanese quail (XP_015714194), house mouse (AAZ38894.1), and chicken (AAZ38896.1).

Figure┬Ā3
LIN28B gene expression profile in ten tissues. 1, Ovary; 2, spleen; 3, uterus; 4, kidney; 5, hypothalamus; 6, pituitary; 7, lung; 8, heart; 9, liver; 10, oviduct; M, DL2000 Marker. LIN28B, lin-28 homolog B; ACTB, beta-actin.
Figure┬Ā4
LIN28B relative mRNA levels in the hypothalamus, pituitary and ovary. Different letters above the bars in the same tissue indicate significant differences (p<0.05). LIN28B, lin-28 homolog B.
Figure┬Ā5
LIN28B relative protein levels in the hypothalamus, pituitary and ovary. Different letters above the bars in the same tissue indicate significant differences (p<0.05). LIN28B, lin-28 homolog B.

Figure┬Ā6
LIN28B protein expression in hypothalamus, pituitary and ovary of preubertal sheep. LIN28B, lin-28 homolog B.

Table┬Ā1
Primers used for LIN28B gene cloning
REFERENCES
1. Nonneman DJ, Schneider JF, Lents CA, et al. Genome wide Association and identification of candidate genes for age at puberty in swine. BMC Genet 20617:50



2. Xin WS, Zhang F, Yan GR, et al. A whole genome sequence association study for puberty in a large Duroc├ŚErhualian F2 population. Anim Genet 2018; 19:29ŌĆō35.

3. Roth CL, Ojeda SR. Genes involved in the neuroendocrine control of normal puberty and abnormal puberty of central origin. Pediatr Endocrinol Rev 2005; 3:67ŌĆō76.

4. Gajdos ZK, Henderson KD, Hirschhorn JN, Palmert MR. Genetic determinants of pubertal timing in the general population. Mol Cell Endocrinol 2010; 324:21ŌĆō9.



5. He C, Kraft P, Chen C, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet 2009; 41:724ŌĆō8.




6. Perry JR, Stolk L, Franceschini N. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet 2009; 41:648ŌĆō50.




7. Elks CE, Perry JR, Sulem P, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet 2010; 42:1077ŌĆō85.




8. Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol 2003; 258:432ŌĆō42.


10. Guo Y, Chen Y, Ito H, et al. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 2006; 384:51ŌĆō61.


11. Slack FJ, Basson M, Liu Z, et al. The lin-41 RBCC gene acts in the C. Elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 2000; 5:659ŌĆō69.


12. Gaytan F, Sangiao-Alvarellos S, Manfredi-Lozan M. Distinct expression patterns predict differential roles of the miRNA-binding proteins. Lin28 and LIN28B, in the mouse testis: studies during postnatal development and in a model of hypogonadotropic hypogonadism. Endocrinology 2013; 154:1321ŌĆō36.



13. Grieco A, Rzeczkowska P, Alm C, Palmert MR. Investigation of peripubertal expression of Lin28a and LIN28B in C57BL/6 female mice. Mol Cell Endocrinol 2013; 365:241ŌĆō8.


14. Sangiao-Alvarellos S, Manfredi-Lozano M, et al. Changes in hypothalamic expression of the Lin28/let-7 system and related micro RNAs during postnatal maturation and after experimental manipulations of puberty. Endocrinology 2013; 154:942ŌĆō55.




15. Sangiao-Alvarellos S, Manfredi-Lozano M, Ruiz-Pino F, et al. Testicular expression of the Lin28/let-7 system: Hormonal regulation and changes during postnatal maturation and after manipulations of puberty. Sci Rep 2015; 5:15683



16. Corre C, Shinoda G, Zhu H, et al. Sex-specific regulation of weight and puberty by the Lin28/let-7 axis. J Endocrinol 2016; 228:179ŌĆō91.


17. Zhu H, Shah S, Shyh-Chang N. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet 2010; 42:626ŌĆō30.




18. Wentong Hou. The important local livestock or poultry genetic resource in the northwest of China. China Agriculture Press; Beijing: 2010.
19. Dantas A, Siqueira ER, Fernandes S, Oba E, Castilho AM. Influence of feeding differentiation on the age at onset of puberty in Brazilian Bergamasca dairy ewe lambs. Arq Bras Med Vet Zootec 2016; 68:22ŌĆō8.


20. Cao GL, Feng T, Chu MX, et al. Subtraction suppressive hybridisation analysis of differentially expressed genes associated with puberty in the goat hypothalamus. Reprod Fertil Dev 2015; 15:10.1071/RD14434

21. Livak KJ, Schmittgen TD. Analysis of relative gene expression dara using real-time quantitative PCR and 2(ŌłÆDelta Delta C(T) method. Methods 2010; 25:402ŌĆō8.

22. Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 2017; 18:437ŌĆō51.



23. Cao G1, Liu Q, Chu M, et al. Analysis on cDNA sequence, alternative splicing and polymorphisms associated with timing of puberty of LIN28B gene in goats. Mol Biol Rep 2013; 40:4675ŌĆō83.


24. Dong N, Liu Y, Zhang T, et al. Different expression patterns of Lin28 and LIN28B in mouse molar development. Arch Oral Biol 2017; 82:280ŌĆō5.


25. Zhang YL. The puberty development and expression of LIN28B-let-7 in the reproductive axis of Jining Qingshan sheep. masterŌĆÖs thesis. Liaocheng, China: Liaocheng University; 2016.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





