 |
 |
| Anim Biosci > Volume 33(12); 2020 > Article |
|
Abstract
Objective
The purpose of this study was to reveal the metabolic shift in the fungus co-cultured with the methanogen (Methanobrevibacter thaueri).
Methods
Gas chromatography-mass spectrometry was used to investigate the metabolites in anaerobic fungal (Pecoramyces sp. F1) cells and the supernatant.
Results
A total of 104 and 102 metabolites were detected in the fungal cells and the supernatant, respectively. The partial least squares-discriminant analysis showed that the metabolite profiles in both the fungal cell and the supernatant were distinctly shifted when co-cultured with methanogen. Statistically, 16 and 30 metabolites were significantly (p<0.05) affected in the fungal cell and the supernatant, respectively by the co-cultured methanogen. Metabolic pathway analysis showed that co-culturing with methanogen reduced the production of lactate from pyruvate in the cytosol and increased metabolism in the hydrogenosomes of the anaerobic fungus. Citrate was accumulated in the cytosol of the fungus co-cultured with the methanogen.
Conclusion
The co-culture of the anaerobic fungus and the methanogen is a good model for studying the microbial interaction between H2-producing and H2-utilizing microorganisms. However, metabolism in hydrogenosome needs to be further studied to gain better insight in the hydrogen transfer among microorganisms.
Anaerobic fungi, found in the rumen, hindgut, and feces of ruminants and other herbivores, are important fiber degrading microoganisms [1]. They are assigned to the phylum Neocallimastigomycota and 11 genera have been described so far, namely: Neocallimastix, Caecomyces, Piromyces, Orpinomyces, Anaeromyces, Cyllamyces, Buwchfawromyces, Oontomyces, Pecoramyces, Feramyces, and Liebetanzomyces [2]. Anaerobic fungi not only produce a variety of fiber degrading enzymes, but also penetrate the plant structure by their rhizoids [3].
Previous studies showed that the fiber degrading ability of anaerobic fungi was significantly improved by co-cultured H2-utilizing methanogens [4–6]. This improvement might be due to shift in the metabolic pathways in anaerobic fungi, which increases production of substrates for the co-cultured methanogens and thus reduces production of other metabolites. Bauchop & Mountfort cultivated an anaerobic fungus, Neocallimastix frontalis, with H2-formate-utilizing methanogen and found that concentrations of lactate, ethanol, and formate were significantly decreased, while the concentration of acetate was significantly increased [5]. During the last decades, similar results have been reported in several studies [6–8].
Using genomic, transcriptomic, and proteomic data, Li et al investigated the shift in the metabolic pathways of anaerobic fungus Pecoramyces sp. F1 co-cultured with a syntrophic methanogen [2]. These workers demonstrated that the expression of many genes involved in the metabolic pathway was downregulated by the syntrophic methanogen. However, only a limited number of proteins involved in the metabolic pathway of the anaerobic fungus were downregulated and no significant positive correlation was observed between the transcriptomic data and the proteomic data. The purpose of the study was to investigate the metabolites in an anaerobic fungus (Pecoramyces sp. F1) and the metabolic shift that occurs when it is co-cultured with a methanogen (Methanobrevibacter thaueri).
The co-culture of anaerobic fungus and methanogen used in the present study was isolated from the rumen of goat [6]. The collection procedure of rumen fluid was approved by the Animals Care and Use Committee of Nanjing Agricultural University (SYXK (SU) 2017–0007). The anaerobic fungus was Pecoramyces sp. F1 [2] and the methanogen was Methanobrevibacter thaueri [6]. The co-culture was maintained in a medium containing rumen fluid with 1% (w/v) rice straw as a substrate and transferred every 3 days in the fresh medium [8]. The culture medium contained penicillin (1,600 U/mL) and streptomycin (2,000 U/mL) to inhibit growth of bacteria.
The monoculture of the anaerobic fungus was obtained by adding chloramphenicol at a final concentration of 50 mg/L, which inhibits growth of methanogens and bacteria in the co-culture [8]. The monoculture was transferred every 3 days in the same medium as used for the co-culture.
For the experiment, a modified medium M2, without rumen fluid, was used with 20 mM glucose as the substrate [9]. The medium (90 mL) after adjusting pH to 6.8 was anaerobically dispensed into a serum bottle (180 mL capacity). The serum bottle with 100% CO2 headspace was sealed with butyl rubber stopper and aluminum crimp to maintain anaerobic condition [10]. Ten bottles were used for mono- and co-cultures, respectively (i.e. a total of 20 bottles), with 5 replicates each for the experimental group and the control group. The experimental group was inoculated with 10 mL of the 3-day old culture and the control group was inoculated with 10 mL of the blank medium. Thereafter, the bottles were incubated at 39°C for 72 h without shaking [8]. After inoculation, the air pressure in the fermentation bottle was balanced with a pressure converter to make the initial air pressure 0. At the end of the fermentation, samples of the supernatant and the fungal cells were collected according to the methods of Marvin-Sikkema et al [11]. The bottles were centrifuged at 200 g for 10 min to separate the fungal cells and the supernatant. The fungal cells were washed using 20 mM phosphate-buffered saline (PBS) containing 20 mM dithiothreitol and centrifugated at 200 g for 10 min. The procedure was repeated twice, and the pellet was dissolved in 2 mL PBS. To break the cells, 1 mm glass beads were added, and the mixture was vortexed for 10 min. It was then centrifuged at 500 g for 15 min. The supernatant was collected to obtain cell contents. The fungal supernatant was centrifuged at 13,000 g for 10 min before the analysis. The samples were stored at −80°C for the gas chromatography-mass spectrometry (GC/MS) analysis.
To an aliquot of 50 μL of the cell content or the fungal supernatant was added 200 μL methanol containing 12.5 μg/mL myristic-1,2-13C2 acid as an internal standard (IS). The mixture was vortexed for 5 min, placed at 4°C for 1 h, and centrifuged at 20,000 g for 10 min at 4°C. An aliquot of 100 μL supernatant was transferred into a GC vial and evaporated to dryness under vacuum (Thermo Fisher Scientific, Asheville, NC, USA). An aliquot of 30 μL methoxyamine in pyridine (10 mg/mL) was added and vortexed for 3 min. After 16 h of methoximation reaction at room temperature, 30 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide containing 1% trimethylchlorosilane was added, vortexed for 1 min, and placed at room temperature for 1 h for trimethylsilylation. Finally, 30 μL methyl myristate in heptane (30 μg/mL) was added and vortexed, and an aliquot of 0.5 μL was used for the GC/MS analysis.
The analysis of metabolites was conducted by a GC/MS system (SHIMADZU GC/MS QP2010Ultra/SE, Kyoto, Japan) fitted with a RTx-5MS capillary column (30 m×0.25 mm, Agilent J&W Scientific, Folsom, CA, USA). The conditions used for GC/MS analysis were as follows: Helium as carrier gas at flow rate of 1.5 mL/min; initial column temperature of 70°C for 3 min followed by increasing from 70°C to 300°C at a rate of 20°C/min and held for 3 min; transfer line temperature of 205°C and ion source temperature of 200°C; ion source voltage of −70 eV and acceleration voltage of −950 V; and solvent delay of 300 s. The MS data were acquired over the range between m/z 50 – 800 [12].
After rejecting the peaks with signal-to-noise (S/N) lower than 30 and normalizing with IS, the retention time, retention index, and mass spectra of each peak were obtained and compared with the reference standards and database including NIST library (2008), Wiley library, and in-house database established by China Pharmaceutical University [12,13]. The partial least squares-discriminant analysis (PLS-DA) was conducted using the R package ropls [14] and significant difference between two groups was considered at p<0.05 and variable importance plot (VIP)>1. Data visualization was done in R studio (www.rstudio.com).
The typical GC/MS chromatograms of metabolites in cells of the anaerobic fungus with/without the methanogen are shown in Figure 1. A total of 104 metabolites were detected in the fungal cell and a distinct difference of the metabolite profiles between the two groups was observed in the PLS-DA score plot (Figure 2A). For evaluation of the model, the R2X, R2Y, and Q2 (cum) were calculated as 0.532, 0.997, and 0.886, respectively. Statistical analysis showed that 16 metabolites differed significantly between the two groups (p<0.05) (Figure 2B). As shown in Table 1, 8 metabolites significantly increased in the co-culture of the anaerobic fungus and the methanogen including 6-hydroxy-9H-purine, citric acid, threonine, glutamic acid, isoleucine, monostearin, uracil and valine. The remaining 8 metabolites were significantly decreased, including D-galactono-1,4-lactone, glucose, ribopyranose, beta-D-methylglucopyranoside, ribonic acid and three unknown metabolites. Many of these amino acids participate in cytosol metabolism, and citric acid is an important intermediate in the oxidation pathway of the tricarboxylic acid (TCA) cycle in cytosol metabolism.
The typical GC/MS chromatograms of metabolites in supernatant of the anaerobic fungus with/without the methanogen are shown in Figure 3. A total of 102 metabolites were detected in the supernatant of the cultures and a distinct difference in the metabolite profiles between the two groups was observed in the PLS-DA score plot (Figure 4A). The R2X, R2Y, and Q2 (cum) of the model were 0.453, 0.99, and 0.898, respectively. Statistical analysis showed that 30 metabolites differed significantly between the two groups (Figure 4B). As shown in Table 2, 22 metabolites were significantly increased in the co-culture of the anaerobic fungus and the methanogen and the remaining 8 metabolites were significantly decreased.
Based on the metabolite profiles in the cells of the anaerobic fungus with/without the co-cultured methanogen and previous reports [2,15], the metabolic pathway of the anaerobic fungus is shown in Figure 5. Combined with the previous research conducted in our laboratory, including the study of the metabolic pathway differences in the utilization of glucose by Pecoramyces sp. F1 in monoculture and co-culture with the methanogen, as well as the research on the effect of the co-cultured methanogen on the dynamic profile of intermediate and end metabolic pathway of Pecoramyces sp. F1, we conclude that co-cultured methanogen stimulated the metabolic pathway to citrate and malate in the cytosol of the anaerobic fungus and stimulated the metabolism in its hydrogenosome.
In the recent past, several studies have focused on the effects of culturing anaerobic fungi with methanogens. For example, several workers [4–7,16] have shown higher fiber degrading ability of anaerobic fungi when co-cultured with methanogens as compared with when they were grown as a monoculture. So far, approximately 60 co-cultures of anaerobic fungi and methanogen combinations have been isolated from different herbivores [6,17–20]. The co-culture of Pecoramyces sp. F1 and Methanobrevibacter thaueri, used in the present study, is one of such co-cultures, which were isolated from the rumen of Chinese local goat and demonstrated to have higher fiber degrading ability than others [6]. The anaerobic fungus in the co-culture was identified as Piromyces sp. F1 according to its morphology and renamed as Pecoramyces sp. F1 by using molecular techniques [2].
Present study showed that the metabolic pathways in the anaerobic fungal cell were shifted by the co-cultured methanogen. The pathways in the cytosol were inhibited and the pathways in the hydrogenosome were stimulated, resulting in increased provision of H+ for the co-cultured methanogen. The production of lactate from pyruvate in the cytosol decreased in the supernatant by 15%, which is a process of H+ consumption (from nicotinamide adenine dinucleotide to nicotinamide adenosine denucleotide) [8,15,21]. The anaerobic fungus does not possess a complete TCA cycle in the cytosol but has reductive (from oxaloacetate to succinate) and oxidative (from citrate to α-ketoglutarate) branches of it [22,23]. Present study showed that citrate was accumulated in the fungal cytosol when co-cultured with the methanogen, which was revealed by the increase of citrate in the fungal cell (7.62-fold). This is consistent with our previous report that citrate was accumulated in the co-culture of the anaerobic fungus and the methanogen [22]. However, no observed (p = 0.265) increase of α-ketoglutarate in the cytosol of the anaerobic fungus co-cultured with the methanogen implies that the oxidative branch was not affected by the co-cultured methanogen, except for citrate accumulation. The content of fumarate in the fungal cells between two groups was not significantly different (p = 1), and even an increase in malate was observed (3.305-fold in the cell, p = 0.056).
Previous studies reported significant accumulation of acetate in the supernatant of the anaerobic fungus co-cultured with the methanogen [6–8], which implied that the metabolism in the hydrogenosomes of the anaerobic fungus was stimulated by the co-cultured methanogen. This was proved in the present study by increase in the contents of malate and pyruvate observed in the fungal cell, also the content of malate in the anaerobic fungal cell was lower than that in the anaerobic fungal cell co-cultured with the methanogen, which indicated increased malate availability in the reductive pathway of the TCA cycle in presence of the methanogen, and enhanced reductive pathway in the cytosol of the anaerobic fungus. A higher amount of malate entered the hydrogenosomes and increased formate and H2 production for utilization by the methanogen, thus promoting the metabolism in the hydrogenosomes. Hydrogenosomes are membrane-bound organelles, which have been found in a variety of unicellular anaerobic eukaryotes such as Trichomonas vaginalis, Psalteriomonas lantern, and Neocallimastix [24,25]. Hydrogenosomes are notable for the energy metabolism of anaerobic eukaryotes. They cannot use oxygen but reduce protons to H2, which can be used by co-cultured methanogens to produce methane instantly. The stimulation of the metabolism in the hydrogenosome implies higher H2 production for the co-cultured methanogen, which benefits both the anaerobic fungus (reduction in growth inhibition from fermentation end products) and the co-cultured methanogen (provision of increased amounts of growth substrate). However, the H2 transfer between the anaerobic fungus and the co-cultured methanogen needs to be further studied to obtain better insight into the metabolic interaction among microorganisms.
The co-cultured methanogen reduced the production of lactate from pyruvate in cytosol of the anaerobic fungus and stimulated metabolism in hydrogenosome of the anaerobic fungus. Citrate was accumulated in the fungal cytosol co-cultured with the methanogen. The co-culture of anaerobic fungi and methanogens is a good model for studying the microbial interaction between H2-producing and H2-utilizing microorganisms. However, the hydrogenosome and its metabolism need to be further studied to evaluate the hydrogen transfer among microorganisms.
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (31772627), the “One Belt and One Road” Technical Cooperation Project of Jiangsu Province (BZ2018055), and the Fundamental Research Funds for the Central Universities (KYDK201701).
Figure 1
Typical gas chromatography-mass spectrometry (GC/MS) chromatograms of metabolites in the fungal cells with/without the co-cultured methanogen. (A) Monoculture of the anaerobic fungus. (B) Co-culture of the anaerobic fungus and the methanogen. 1, lactate; 2, alanine; 3, Ua3; 4, oxalate; 5, α-aminobutyric acid; 6, 2-keto-3-methylvaleric acid; 7, valine; 8, carbamic acid; 9, leucine; 10, glycerol-3TMS; 11, isoleucine; 12, fumaric acid; 13, threonine; 14, UN01; 15, lauric acid; 16, taurine; 17, internal standard; 18, histidine; 19, glucose; 20, tyrosine; 21, palmitic acid; 22, oleic acid; 23, stearic acid; 24, UI22; 25, UI23.
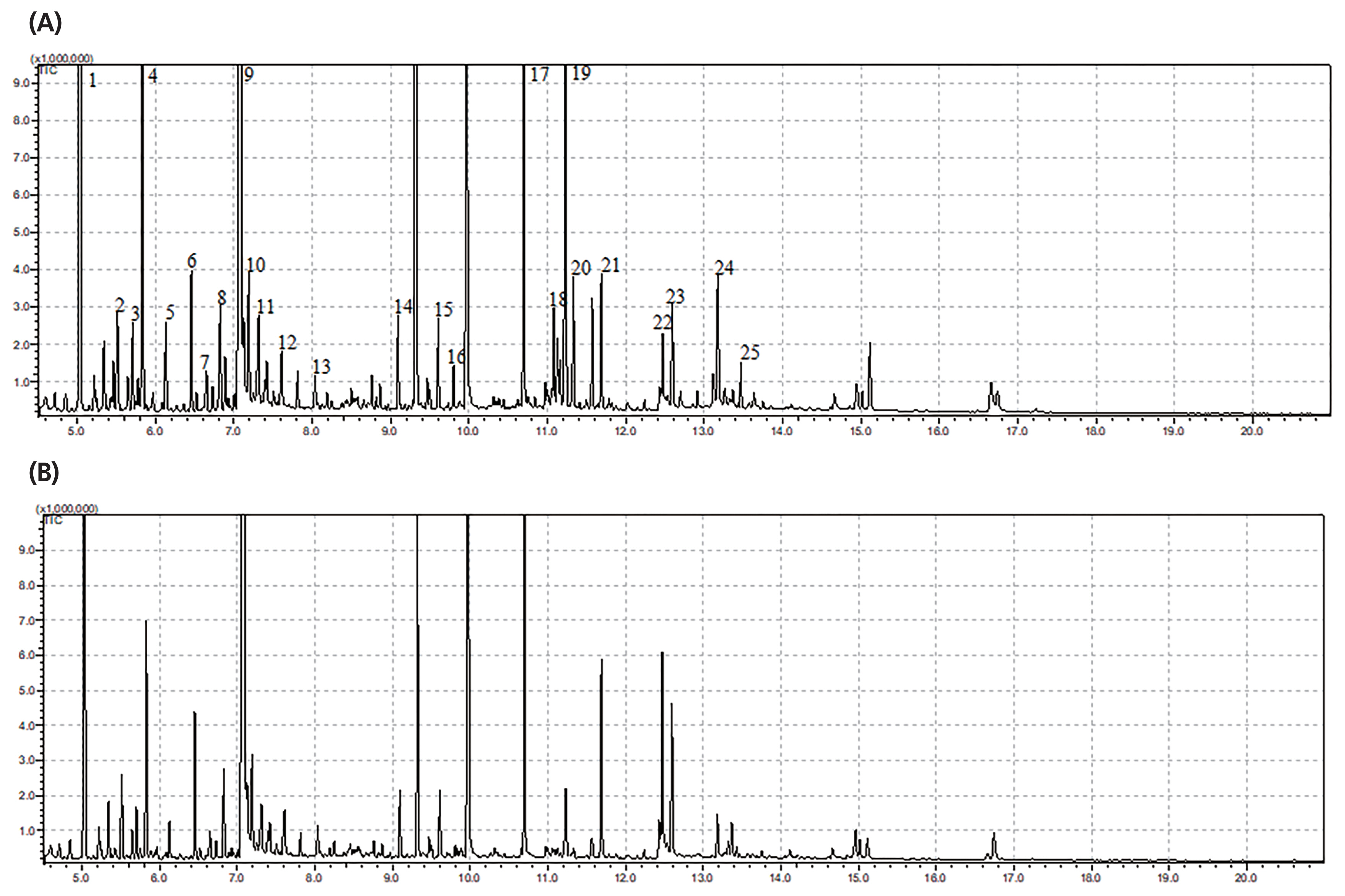
Figure 2
Effects of the co-cultured methanogen on the metabolites in the fungal cell, as evaluated by the PLS-DA analysis. (A) PLS-DA score plot. (B) Relationship between VIP and p-value. PLS-DA, partial least squares-discriminant analysis; VIP, variable importance plot.

Figure 3
Typical gas chromatography-mass spectrometry chromatograms of metabolites in the supernatant of the anaerobic fungus with/without the co-cultured methanogen. (A) Monoculture of anaerobic fungus. (B) Co-culture of anaerobic fungus and methanogen. 1, pyruvic acid; 2, lactate; 3, alanine; 4, oxalate; 5, urea; 6, isoleucine; 7, serine; 8, threonine; 9, aminomalonic acid; 10, malic acid; 11, glutamic acid; 12, taurine; 13, glycerol-3-phosphate; 14, internal standard; 15, glucose; 16, linoleic acid; 17, cystin.

Figure 4
Effects of co-cultured methanogen on the metabolites in the supernatant of cultures as evaluated by PLS-DA analysis. (A) PLS-DA score plot. (B) Relationship between VIP and p-value. PLS-DA, partial least squares-discriminant analysis; VIP, variable importance plot.
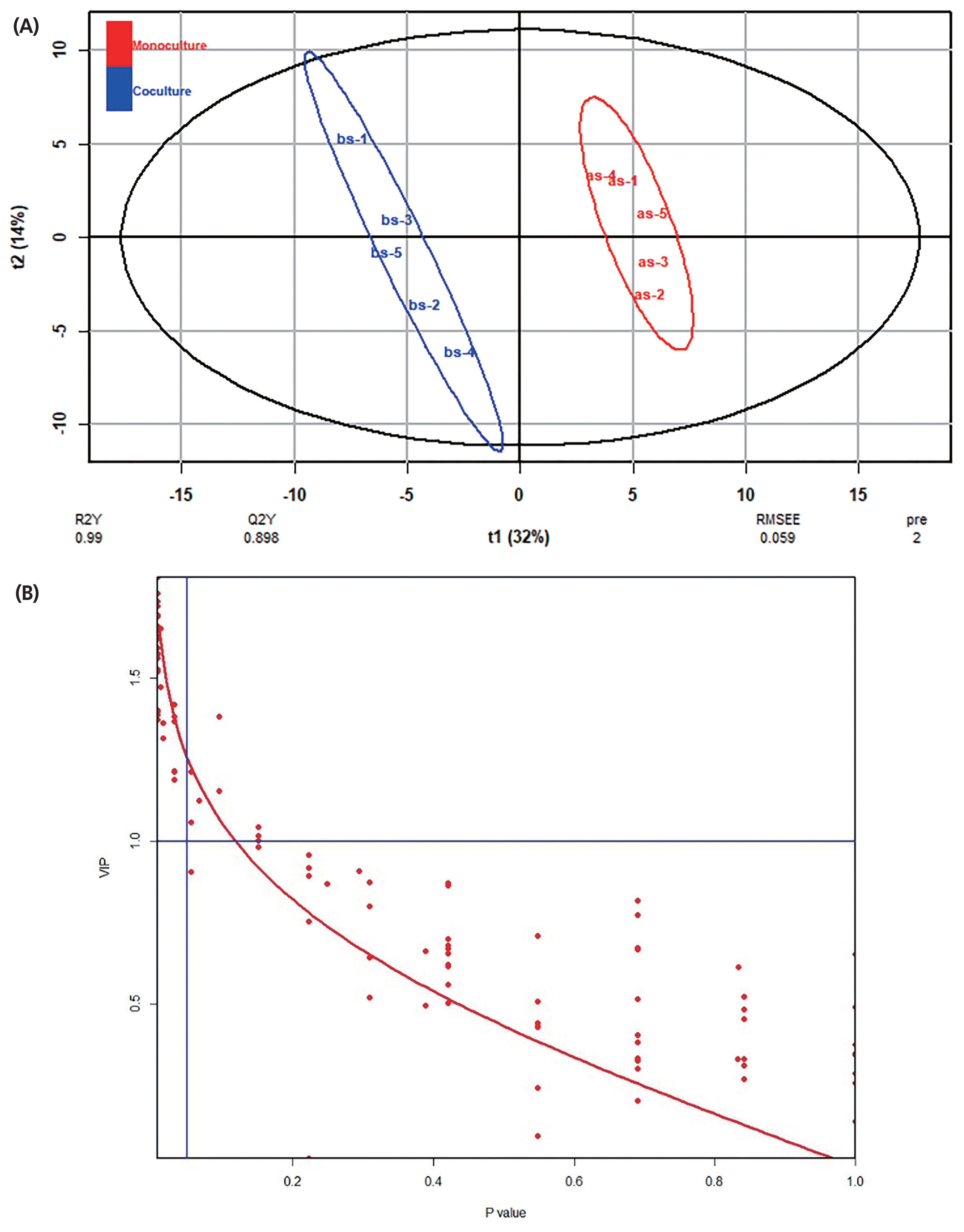
Table 1
Significantly changed metabolites in the anaerobic fungal cells by the co-cultured methanogen
| Compounds | Fold change (bc/ac) | p-value1) | VIP1) |
|---|---|---|---|
| 6-Hydroxy-9H-purine | 2.432 | 0.008 | 1.633 |
| Citric acid | 7.620 | 0.008 | 2.053 |
| D-Galactono-1,4-lactone | 0.189 | 0.008 | 1.320 |
| Glucose | 0.092 | 0.008 | 1.470 |
| Ribopyranose | 0.042 | 0.008 | 1.182 |
| S07 | 0.139 | 0.008 | 1.195 |
| Beta-D-Methylglucopyranoside | 0.094 | 0.012 | 1.524 |
| Threonine | 1.443 | 0.016 | 1.547 |
| UI23 | 0.298 | 0.016 | 1.660 |
| Glutamic acid | 1.963 | 0.032 | 1.365 |
| Isoleucine | 1.680 | 0.032 | 1.512 |
| Monostearin | 1.497 | 0.032 | 1.342 |
| S04 | 0.654 | 0.032 | 1.241 |
| Uracil | 1.869 | 0.032 | 1.480 |
| Valine | 1.870 | 0.032 | 1.462 |
| Ribonic acid | 0.133 | 0.045 | 1.516 |
Table 2
Significantly changed metabolites in supernatant of the anaerobic fungus by the co-cultured methanogen
| Compounds | Fold change (bs/as) | p-value1) | VIP1) |
|---|---|---|---|
| 6-Hydroxy-9H-purine | 0.575 | 0.008 | 1.688 |
| Alanine | 1.334 | 0.008 | 1.692 |
| Carbamic acid | 1.323 | 0.008 | 1.396 |
| Cystin | 0.105 | 0.008 | 1.809 |
| Fructose | 0.157 | 0.008 | 1.627 |
| Glutamic acid | 1.235 | 0.008 | 1.562 |
| Glutamine | 1.503 | 0.008 | 1.573 |
| Glycerol-3-phosphate | 0.812 | 0.008 | 1.593 |
| Isoleucine | 1.209 | 0.008 | 1.577 |
| Lactate | 0.851 | 0.008 | 1.620 |
| Lauric acid | 1.371 | 0.008 | 1.527 |
| Malic acid | 0.273 | 0.008 | 1.736 |
| Myo-inositol | 2.534 | 0.008 | 1.660 |
| Myo-inositol-2-phosphate | 1.767 | 0.008 | 1.372 |
| Oxalate | 2.195 | 0.008 | 1.387 |
| Proline | 1.763 | 0.008 | 1.403 |
| Pyruvic acid | 2.655 | 0.008 | 1.695 |
| Threonine | 1.293 | 0.008 | 1.761 |
| UN01 | 2.983 | 0.008 | 1.521 |
| Valine | 1.387 | 0.008 | 1.722 |
| Citric acid | 0.213 | 0.012 | 1.653 |
| Ethanedioic acid | 2.913 | 0.012 | 1.473 |
| Glycine | 1.245 | 0.016 | 1.363 |
| Heptanoic acid | 1.132 | 0.016 | 1.316 |
| 9-Octadecenoic-acid | 2.926 | 0.032 | 1.212 |
| Fumaric acid | 1.269 | 0.032 | 1.382 |
| Glycerol-3TMS | 1.180 | 0.032 | 1.593 |
| L-Asparagine | 1.209 | 0.032 | 1.419 |
| Ua8 | 0.487 | 0.032 | 1.188 |
| Uracil | 1.266 | 0.032 | 1.367 |
REFERENCES
1. Orpin CG. Studies on the rumen flagellate Neocallimastix frontalis
. J Gen Microbiol 1975; 91:249–62.
https://doi.org/10.1099/00221287-91-2-249


2. Li Y, Li Y, Jin W, et al. Combined genomic, transcriptomic, proteomic, and physiological characterization of the growth of Pecoramyces sp. F1 in monoculture and co-culture with a syntrophic methanogen. Front Microbiol 2019; 10:435
https://doi.org/10.3389/fmicb.2019.00435



3. Edwards JE, Forster RJ, Callaghan TM, et al. PCR and omics based techniques to study the diversity, ecology and biology of anaerobic fungi: insights, challenges and opportunities. Front Microbiol 2017; 8:1657
https://doi.org/10.3389/fmicb.2017.01657



4. Teunissen MJ, Kets EPW, Op den Camp HJM, Huis in’t Veld JHJ, Vogels GD. Effect of coculture of anaerobic fungi isolated from ruminants and non-ruminants with methanogenic bacteria on cellulolytic and xylanolytic enzyme activities. Arch Microbiol 1992; 157:176–82.
https://doi.org/10.1007/BF00245287



5. Bauchop T, Mountfort DO. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl Environ Microbiol 1981; 42:1103–10.



6. Jin W, Cheng YF, Mao SY, Zhu WY. Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour Technol 2011; 102:7925–31.
https://doi.org/10.1016/j.biortech.2011.06.026


7. Joblin KN, Naylor GE, Williams AG. Effect of Methanobrevibacter smithii on xylanolytic activity of anaerobic ruminal fungi. Appl Environ Microbiol 1990; 56:2287–95.



8. Cheng YF, Edwards JE, Allison GG, Zhu WY, Theodorou MK. Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour Technol 2009; 100:4821–8.
https://doi.org/10.1016/j.biortech.2009.04.031


9. Barichievich EM, Calza RE. Supernatant protein and cellulase activities of the anaerobic ruminal fungus Neocallimastix frontalis EB188. Appl Environ Microbiol 1990; 56:43–8.



10. Haitjema CH, Solomon KV, Henske JK, Theodorou MK, O’Malley MA. Anaerobic gut fungi: Advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol Bioeng 2014; 111:1471–82.
https://doi.org/10.1002/bit.25264


11. Marvin-Sikkema FD, Gomes TMP, Grivet JP, Gottschal JC, Prins RA. Characterization of hydrogenosomes and their role in glucose metabolism of Neocallimastix sp. L2. Arch Microbiol 1993; 160:388–96.
https://doi.org/10.1007/BF00252226


12. Ma B, Liu J, Zhang Q, et al. Metabolomic profiles delineate signature metabolic shifts during estrogen deficiency-induced bone loss in rat by GC-TOF/MS. PLOS One 2013; 8:e54965
https://doi.org/10.1371/journal.pone.0054965



13. Ma B, Li X, Zhang Q, et al. Metabonomic profiling in studying anti-osteoporosis effects of strontium fructose 1,6-diphosphate on estrogen deficiency-induced osteoporosis in rats by GC/TOF-MS. Eur J Pharmacol 2013; 718:524–32.
https://doi.org/10.1016/j.ejphar.2013.06.030


14. Thevenot EA, Roux A, Xu Y, Ezan E, Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J Proteome Res 2015; 14:3322–35.
https://doi.org/10.1021/acs.jproteome.5b00354


15. Li Y, Jin W, Cheng Y, Zhu W. Effect of the associated methanogen Methanobrevibacter thaueri on the dynamic profile of end and intermediate metabolites of anaerobic fungus Piromyces sp. F1. Curr Microbiol 2016; 73:434–41.
https://doi.org/10.1007/s00284-016-1078-9



16. Nakashimada Y, Srinivasan K, Murakami M, Nishio N. Direct conversion of cellulose to methane by anaerobic fungus Neocallimastix frontalis and defined methanogens. Biotechnol Lett 2000; 22:223–7.
https://doi.org/10.1023/A:1005666428494

17. Leis S, Dresch P, Peintner U, et al. Finding a robust strain for biomethanation: anaerobic fungi (Neocallimastigomycota) from the Alpine ibex (Capra ibex) and their associated methanogens. Anaerobe 2014; 29:34–43.
https://doi.org/10.1016/j.anaerobe.2013.12.002


18. Wei YQ, Yang HJ, Luan Y, Long RJ, Wu YJ, Wang ZY. Isolation, identification and fibrolytic characteristics of rumen fungi grown with indigenous methanogen from yaks (Bos grunniens) grazing on the Qinghai-Tibetan Plateau. J Appl Microbiol 2016; 120:571–87.
https://doi.org/10.1111/jam.13035


19. Wei YQ, Long RJ, Yang H, et al. Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens) grazing on the Qinghai Tibetan Plateau. Anaerobe 2016; 39:158–64.
https://doi.org/10.1016/j.anaerobe.2016.03.005


20. Sun M, Jin W, Li Y, Mao S, Cheng Y, Zhu W. Isolation and identification of cellulolytic anaerobic fungi and their associated methanogens from Holstein cow. Acta Microbiologica Sinica 2014; 54:563–71.
https://doi.org/10.13343/j.cnki.wsxb.2014.05.011


21. Li YF, Jin W, Mu CL, Cheng YF, Zhu WY. Indigenously associated methanogens intensified the metabolism in hydrogenosomes of anaerobic fungi with xylose as substrate. J Basic Microbiol 2017; 57:933–40.
https://doi.org/10.1002/jobm.201700132


22. Cheng YF, Jin W, Mao SY, Zhu WY. Production of citrate by anaerobic fungi in the presence of co-culture methanogens as revealed by 1H NMR spectrometry. Asian-Australas J Anim Sci 2013; 26:1416–23.
https://doi.org/10.5713/ajas.2013.13134




23. Kown M, Song JY, Ha JK, Park HS, Chang JS. Analysis of functional genes in carbohydrate metabolic pathway of anaerobic rumen fungus Neocallimastix frontalis PMA02. Asian-Australas J Anim Sci 2009; 22:1555–65.
https://doi.org/10.5713/ajas.2009.80371


24. Hackstein JHP, Akhmanova A, Boxma B, Harhangi HR, Voncken FGJ. Hydrogenosomes: eukaryotic adaptations to anaerobic environments. Trends Microbiol 1999; 7:441–7.
https://doi.org/10.1016/S0966-842X(99)01613-3


25. Akhmanova A, Voncken FGJ, Hosea KM, et al. A hydrogenosome with pyruvate formate-lyase: Anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mol Microbiol 1999; 32:1103–14.
https://doi.org/10.1046/j.1365-2958.1999.01434.x


- TOOLS







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





