Genome-wide Association Study to Identify Quantitative Trait Loci for Meat and Carcass Quality Traits in Berkshire
Article information
Abstract
Meat and carcass quality attributes are of crucial importance influencing consumer preference and profitability in the pork industry. A set of 400 Berkshire pigs were collected from Dasan breeding farm, Namwon, Chonbuk province, Korea that were born between 2012 and 2013. To perform genome wide association studies (GWAS), eleven meat and carcass quality traits were considered, including carcass weight, backfat thickness, pH value after 24 hours (pH24), Commission Internationale de l’Eclairage lightness in meat color (CIE L), redness in meat color (CIE a), yellowness in meat color (CIE b), filtering, drip loss, heat loss, shear force and marbling score. All of the 400 animals were genotyped with the Porcine 62K SNP BeadChips (Illumina Inc., USA). A SAS general linear model procedure (SAS version 9.2) was used to pre-adjust the animal phenotypes before GWAS with sire and sex effects as fixed effects and slaughter age as a covariate. After fitting the fixed and covariate factors in the model, the residuals of the phenotype regressed on additive effects of each single nucleotide polymorphism (SNP) under a linear regression model (PLINK version 1.07). The significant SNPs after permutation testing at a chromosome-wise level were subjected to stepwise regression analysis to determine the best set of SNP markers. A total of 55 significant (p<0.05) SNPs or quantitative trait loci (QTL) were detected on various chromosomes. The QTLs explained from 5.06% to 8.28% of the total phenotypic variation of the traits. Some QTLs with pleiotropic effect were also identified. A pair of significant QTL for pH24 was also found to affect both CIE L and drip loss percentage. The significant QTL after characterization of the functional candidate genes on the QTL or around the QTL region may be effectively and efficiently used in marker assisted selection to achieve enhanced genetic improvement of the trait considered.
INTRODUCTION
Consumer demands for pork depend on various meat quality parameters such as odor, flavor, tenderness and juiciness. Additionally, some physical and biochemical parameters such as pH, shear force (SF), water holding capacity and intramuscular fat content of meat are also important pork quality determinants (Bonneau and Lebret, 2010).
Limitations in measuring carcass traits in live animals have made it difficult to improve the traits through traditional selection approaches (Suzuki et al., 2005). Therefore, the use of molecular genetic tools can overcome these limits, and offer new opportunities for genetic improvement of the traits (Davoli and Braglia, 2008). Recently established genome-wide single nucleotide polymorphism (SNP) panel has enabled the mapping of quantitative trait loci (QTL) and prediction of animal’s genetic merit for the traits of interest (Goddard and Hayes, 2009). A genome-wide association study (GWAS) can significantly improve our understanding of the genetic mechanism underlying the complex traits (Lee et al., 2013). Therefore, the identification of the genomic regions underlying pork quality traits and their applications via marker-assisted selection would allow pig breeders to select at an early age (Luo et al., 2012). GWAS with high density markers exploits linkage disequilibrium (LD) for QTL mapping (Meuwissen et al., 2001).
Berkshire (Susscrofa domesticus) pork possesses some merits of meat quality traits such as richness, texture, marbling, juiciness, tenderness and flavor (Goodwin and Burroughs, 1995). Several GWAS in pigs were reported in many pure and crossbreds for various traits including meat and carcass quality traits (Sanchez et al., 2014). To date, 13,958 QTLs were detected in pigs for 668 different traits (animalgenome.org/cgi-bin/QTLdb/index, Release on September 06, 2015).
The main objectives in this study were to perform GWAS with a high-density Porcine SNP62K BeadChip (Illumina, San Diego, CA, USA) in Berkshire to detect significant SNPs or QTLs associated with meat and carcass quality traits, and to detect candidate genes influencing the traits nearby the detected QTLs.
MATERIALS AND METHODS
Materials and methods
Animals and management
A set of samples (n = 400) from Berkshire pigs that were born between 2012 and 2013 were collected from Dasan breeding farm, Namwon, Cheonbuk province, Korea. The piglets were weaned at 3 to 4 weeks of age and moved into piglet pens, in each of which about 100 piglets were raised for 60 days. Then, the pigs were placed in growth/fattening pens 20 pigs in size for 90 to 120 days. The pigs were fed with the commercial feeds according to the regiments of Purina Ltd.
Carcass and meat quality measurements
The pigs were slaughtered approximately at 211(±23) days of age in an abattoir in Namwon and cooled subsequently at 0ºC for 24 hour in a chilling room. Carcass and meat quality measurements were performed at Sunchon National University, Korea. For each individual, slaughter age (sage), sex, and year-season of birth were also recorded.
Carcass weight (CWT) and backfat thickness (BF) of each carcass were measured. The parts of loins (m. longissimus dorsi) on the left side of the cold carcasses were sampled to determine meat quality parameters. As soon as collected all samples were placed in vacuum bags, transported to the laboratory and then frozen at −50ºC until they were analyzed. For analysis of drip loss, and heat loss, only the subcutaneous fat of meat samples was removed. For the other traits, all visible fat was trimmed off. The surface color and marbling scores of each loin was categorized based on National Pork Producers Council (NPPC) standard (NPCC, 2000). The surface color values were measured by the Commission Internationale de l’Eclairage (CIE) L* (lightness), a* (redness), and b* (yellowness) system using a Minolta colorimeter (Model CR-410, Minolta Co. Ltd., Japan). The colorimeter was calibrated against a white reference tile plate (L* = 89.2, a* = 0.921, b* = 0.783). The averaged value of five random measurements taken from different locations was used for the statistical analysis. The pH value was determined using a pH meter (Orion 2 Star, Thermo scientific, Beverly, MA 01915, USA). The drip loss was measured as the percentage weight loss of a standardized (3×3×3 cm) meat sample placed in a sealed petri-dish at 4ºC during the storage of 2 days. The heat (cooking) loss was determined as the percentage weight loss of a standardized (3×3×3 cm) meat sample after cooking in an electric grill with double pans (Nova EMG-533, 1,400 W, Evergreen enterprise, Seoul, Korea) for 90 s until the internal temperature of the meat sample reached 72ºC. For, SF measurement, the samples were prepared a cubic form (30×30×20 mm), heated until internal temperature of the samples reached 72ºC±2ºC, and then cooled for 30 min at room temperature. Each sample was cut perpendicular to the longitudinal orientation of the muscle fiber with a Warner-Bratzler shear attachment on a texture analyzer (TA-XT2, Stable Micro System Ltd., Godalming, UK). The maximum SF value (kg) was recorded for each sample. Test and pre-test speeds were set at 2.0 mm/s while the post-test speeds were set at 5.0 mm/s.
Molecular data
Using the Infinium HD Assay Ultra Protocol (Illumina), all of the 400 animals were genotyped with the Porcine 62K SNP BeadChips (Illumina Inc., USA). A total of 62,163 SNPs were embedded on the chip that covered the entire porcine genome. For subsequent data analysis, a subset of SNP was selected by removing the SNPs on sex chromosomes. After initial filtering, 56,517 SNPs were used for further analysis. All SNPs underwent the process of quality control (QC) based on the following criteria: i) eliminating individuals with call rates less than 90%, ii) removing SNPs with minor allele frequency less than 1%, iii) filtering out SNPs with call rates less than 90%, iv) removing SNPs with significant departure from Hardy Weinberg equilibrium (p<0.001). These procedures were implemented with PLINK v1.7 (Purcell et al., 2007). A number of 41,082 SNPs and 313 individuals passing QC thresholds were used for further data analysis. The chromosomal positions referred to the pig genome build 10.2 (Gorenen et al., 2012).
Statistical analysis
A SAS general linear model (GLM) procedure (SAS version 9.2) was used to pre-adjust the animal phenotypes before GWAS testing. Both sire and sex effects were fitted as fixed effects, while slaughter age was considered as a covariate in the SAS GLM model. Then, the residuals of each phenotype were regressed on additive effects of each SNP under a simple linear regression additive model (Purcell et al, 2007), by assigning SNP genotypes of AA, AB and BB with 1, 0, and −1, respectively, reflecting a substitution effect of allele A on allele B.
After linear regression analysis, permutation tests were performed for 1,000 times on each chromosome at 0.05 chromosome-wide level, such that the SNPs were arranged in ascending order of the p values (Purcell et al., 2007). The top 50th SNP on each chromosome were selected for further stepwise regression analysis. Among the significant SNPs, the best set of SNP markers were selected using the stepwise regression procedure (Neter et al., 1990), because some of the significant SNPs would yield redundant information due to LD between closely linked SNPs, i.e a non-random association between alleles of different SNPs. Inclusion and exclusion of each SNP out of the model was determined at 0.001 level (Lee et al., 2013). For significant SNPs, phenotypic variation explained by each (jth) SNP (S2snpj ) was calculated as
RESULTS AND DISCUSSIONS
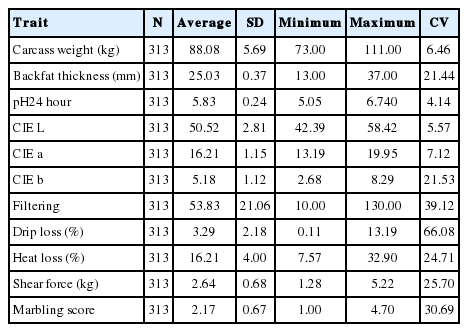
From the summary statistics of the phenotypes of the studied traits, the highest variability of 66.08% was found in drip loss (%), followed by filtering (39.12%), marbling scores (30.69%), SF (25.70%), heat loss (24.71%), CIE b (21.53%), CIE a (7.12%), CIE L (5.57%) and pH 24 hour postmortem (4.14%). The lowest variability was found as 4.14% in pH 24 hour (Table 1).
A total of 55 SNPs were detected on different chromosomes after the stepwise regression procedures for the meat and carcass quality traits. All the SNPs were found to be distributed across almost the entire genome. Some QTL with pleiotropic effects were also identified. A QTL located on Sus scrofa chromosome (SSC)12 at 1.16 Mb affecting pH24 hours postmortem was found to affect pork color L and the QTL on SSC16 at 76.4 Mb for pH24 influenced drip loss percentage (Table 2).

Identities, positions of the SNPs with significant effects on meat quality traits in a Berkshire population
Carcass weight
For CWT, four significant SNPs were detected on SSCs 12, 15, 16, and 17. The SNP located on SSC 17 at 53.4 Mb accounted for the greatest proportion of phenotypic variance of 6.25% to the total variance explained by all SNPs (Table 2). We found a significant SNP at 45.2 Mb on SSC16 for CWT which lies close to the significant SNP reported to be associated with ham weight in commercial Large white pigs by Sanchez et al., 2014). In this study, we detected QTL at 10.78 Mb on SSC12 affecting CWT residing within the genomic region affecting carcass length in a Duroc and Pietrain crossbred population reported by Liu et al. (2007), which supports our findings. The significant SNP at 53.4Mb on SSC17 was located within the significant QTL region (45 to 68.9 Mb) influencing carcass length in Wild Boar, Meishan and Pietrain crossbred population (Pierzchala et al., 2003).
Back fat thickness
Seven significant SNPs (QTLs) for BF were detected, among which the most significant SNP was found at 86.9 Mb on SSC13. On SSCs 13 and 15, a set of three SNPs were found on each chromosome (Table 2). The greatest proportion of phenotypic variance was estimated as 6.93%, explained by the SNP residing at 90.8 Mb of SSC15 (Table 2). We found a cluster of three significant SNPs for backfat located at 55.4 to 101 Mb on SSC15, which agrees with the results of Liu et al. (2007) who detected a QTL for the same trait within the same position. In this study, detection of a significant QTL at 56.3 Mb of SSC16 is supported by Liu et al. (2007) and Edwards et al. (2008) who reported QTL on the same chromosome at 25.8 cM (corresponding to 1.16 to 67.6 Mb) and 55 cM (22.3 to 71.8 Mb) for BF, respectively. Our detected significant SNP on SSC15 at 101.8 Mb is in line with the detected QTL for the backfat at 102 Mb in Italian Large White pig reported by Fontanesi et al. (2012).
pH 24 hours postmortem
We found seven significant SNPs for pH24 on SSCs 1, 6, 12, 13, 16, 17, and 18. The most significant (p<0.001) QTL for pH24 was located at 1.2 Mb position of SSC12, with 8.3% proportion of phenotypic variance (Table 2). In this study, we detected QTL for pH24 at 89.2 Mb of SSC6 which is in agreement with Yue et al. (2003) who reported QTL for the same trait in Wild Boar, Mehisan and Pietrian crosses. A significant SNP at 76.4 Mb of SSC16 underlying pH24 detected in this study is strongly supported by Edwards et al. (2008) who detected a QTL within the similar position in a Duroc-Pietrain crossbred population. Our detected QTL for pH24 on SSC13 at 139.6 Mb lies nearby the QTLS discovered by Evans et al. (2004) affecting pH for Semimembranosus and pH for Longissmus at 62.2 to 79.3 cM (corresponding 89.5 to 194.99 Mb) in a commercial outbred pig.
CIE L
Six QTLs for CIE L were detected on SSCs 1, 6, 7, 8, 9 and 12, among which the QTL at 6.12 Mb of SSC9 accounted for the greatest proportion of phenotypic variance (6.10%) of the total phenotypic variance (Table 2). The QTL at 66 Mb of SSC6 resided very closely to the QTL detected in purebred Duroc pigs by Li et al. (2010). We detected a QTL on SSC1 at 8.2 Mb, while a QTL for color L was reported at the similar position (Sanchez et al., 2014). In this study, a QTL was identified at 6.1 Mb on SSC9 affecting pork color L, which agrees with the discovery a QTL affecting pork color L was in Large White pigs reported by Sanchez et al. (2014). Our findings of a significant SNP at 16.9 Mb on SSC8 is in line with the report of Ma et al. (2009) who also found a QTL for meat color within the same region.
CIE a
Five significant SNPs were detected on SSCs 3, 5, 6, 9 and 11 for redness of pork color (CIE a), among which the QTL at 81.5 Mb of SSC6 explained the greatest proportion of variance of 7.2% (Table 2). Our finding of a QTL on SSC5 at 13.5 Mb is supported by Li et al. (2010) who detected a QTL underlying pork color in the similar region.
CIE b
Seven QTLs were detected for CIE b on SSCs 1, 4, 7, 9, 11 and 17, among which two QTLs were located on SSC4. The QTL at 30.1 Mb on SSC7 explained the greatest proportion of phenotypic variance of 7.1% (Table 2). Our detected QTL for CIE b at 111.8 Mb on SSC4 resided within the QTL region affecting Japanese color scores (Van Wijk et al., 2007). Ponsuksili et al. (2014) reported a QTL for CIE b at 114.7 Mb on SSC4 which is located very close (111.8 Mb) to our detected significant SNP on SSC 4.
Filtering
Two significant SNPs for filtering were found at 7 Mb and 141.3 Mb of SSCs 1 and 9, respectively. Each of the two SNPs explained the same proportion of the phenotypic variance of 6% (Table 2).
Drip loss
We identified two QTLs affecting drip loss percentage in pork on SSCs 3 and 16 at 31.9 Mb and 76.4 Mb, respectively (Table 2). At the region of the QTL on SSC3, Liu et al. (2007) reported a QTL for drip loss in Duroc-Pietrain crossbred pigs.
Heat loss
Seven QTLs for heat loss were detected on SSCs 3, 8, 12, 13, 14, and 15, among which two QTL were located on SSC15 at 953 kb and 1.2 Mb, respectively (Table 2). The greatest proportion of variance (8.2%) was accounted for by the SNP at 32.4 Mb of SSC3 (Table 2). We detected a significant SNP at 0.95 Mb on SSC15 which agrees with the detection of a QTL for heat loss by Rohrer et al. (2006) lying close to our detected significant SNP.
Shear force
Two QTLs for SF was detected on SSC18 at 28.3 Mb and 52.8 Mb, respectively. The greatest proportion of phenotypic variance (5.8%) of the total variance was explained by the QTL at 28.3 Mb (Table 2).
Marbling score
Six significant QTLs for marbling score in pork were found on SSCs 7, 9, 12, 14, and 16. We found two QTLs on SSC12 affecting marbling at 53.9 Mb and at 54.7 Mb which is similar to the report of Luo et al. (2012) who identified significant SNPs in the similar position in Large White-Minzhu and Duroc-Pietrain crossbred pigs.
Candidate genes
According to the functional biological role of genes, candidate genes were searched within 500 kb downstream/upstream of the detected SNPs. Noticeably; five candidate genes were identified nearby the significant SNPs for meat and carcass quality traits (Table 3). Gene annotation was done using Sus scrofa (pig) genome build 9.2. For the two significant SNPs on SSCs 12 and 16 associated with CWT, we found two candidate genes namely protein kinase C, Alpha (PRKCA) and survival of motor neuron (SMN). The PRKCA gene has positive role in mitotic cell cycle, negative regulation of cell proliferation (www.ncbi.nlm.nih.gov) hence affects growth and CWT in pigs. The SMN1 gene located on SSC16 nearby the significant SNP affecting CWT in Berkshire is known to cause skeletal muscle wasting in pigs when it is deleted or mutated (Jeffrey and Randall, 2011) thereby has influence in pig CWT. Two candidate genes myxovirus resistance 1 (MX1) and myxovirus resistance 2 (MX2) were found nearby the significant QTL affecting BF in Berkshire on SSC13. The gene, MX1, contributes to meat quality attributes such as marbling in pig (Feng et al., 2012), while the gene MX2, affects rib fat in pig (Fowler et al., 2013). Moreover, the gene solute carrier family 24 member 5 (SLC24A5), located nearby the identified QTL for pork color (CIE b) on SSC1, is found to affect pigmentation (Rambow et al., 2008) thereby might have some influence on pork color.
CONCLUSION
In this study, 55 significant SNPs (QTL) for pork quality were detected in a Berkshire population. Among the QTL, two QTL on SSC12 and SSC16 for pH24 post mortem were also found to be involved in pork color and drip loss, respectively, indicating a pleiotropic effect. Four candidate genes based on biochemical and physiological roles relevant to meat and carcass quality traits were discovered within 1 Mb distant to the significant markers. The detected QTL need to be validated in other Berkshire population to be used as potential markers via marker-assisted selection programs, which will improve meat and carcass quality traits in a more effective and efficient way than traditional selection and breeding schemes.
ACKNOWLEDGMENTS
This research was supported by a grant (PJ01108301) from the Next Generation BioGreen 21 Program, Rural Development Administration, Republic of Korea.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.