 |
 |
| Anim Biosci > Volume 30(5); 2017 > Article |
|
Abstract
Objective
Experiments were conducted to clone the sequence of Wild Argali short palate, lung and nasal epithelium clone 1 (SPLUNC1) cDNA, and to lay the foundation for further study the biological function of Wild Argali SPLUNC1.
Methods
The complete sequence of Wild Argali SPLUNC1 cDNA was generated by rapid amplification of cDNA ends. The entire coding sequence was inserted into the pPIC9K vector and expressed in Pichia pastoris (P. pastoris) GS115. The recombinant SPLUNC1 protein was detected by Western blot and purified by Ni2+ chelate affinity chromatography. The test of effect of the protein on Mycoplasma ovipneumoniae (MO) was performed with real-time polymerase chain reaction.
Results
The Wild Argali SPLUNC1 cDNA was 1,076 bp with an open reading frame of 768 bp, which encoded a 26.49 kDa protein composed of 255 amino acids. Its amino acid sequence shared 98.4%, 96.9%, 94.5%, 90.2%, 80.8%, 78.4%, 78.3%, 72.5%, 72.3%, 68.8% identity with those of SPLUNC1 cDNA from Ovis aries (accession no. NP_001288334.1), Capra hircus (accession no. XP_005688516.1), Pantholops hodgsonii (accession no. XP_005979709.1), Bos taurus (accession no. NP_776851.1), Felis catus (accession no. XP_006929910.1), Homo sapiens (accession no. NP_001230122.1), Sus scrofa (accession no. NP_001005727.1), Chinchilla lanigera (accession no. NP_001269294.1), Mus musculus (accession no. NP_035256.2), and Rattus norvegicus (accession no. NP_742028.1), respectively. The recombinant protein corresponded to the expected molecular mass of 25.47 kDa as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and it was detected in the supernatant of P. pastoris, and it could be purified. The results from the test of inhibition effect of argali recombinant SPLUNC1 protein on MO showed that the product could inhibit MO very well (p<0.01).
Argali (Ovis ammon), also known as the big sheep in Artiodactyla, Ruminantia, Boridae, Caprinae, genus Ovis, belongs to wild animals under the second class state protection [1]. Wild Argali are distributed in the mountains of Eurasia, and also found in Inner Mongolia, Ningxia, Gansu, Xinjiang, Qinghai, and Tibet in China. Wild Argali’s weight is the highest among Caprinae, and the rams weigh up to 200 kg. Wild Argali are up to 1.27 meters tall at the shoulder, have more lean and less fat content, enabling fast growth, less fat, strong adaptability, high disease resistance as the main characteristics [2], and are an excellent genetic resource. It is important to exploit and make full use of the superior characteristics of Wild Argali in Xinjiang.
Short palate, lung and nasal epithelium clone 1 (SPLUNCl) is a secreted protein molecule, which is expressed highly on respiratory epithelium and has a signal peptide. SPLUNC1 is especially expressed in the oral cavity, nasal cavity, nose, pharynx, respiratory tract, nasopharyngeal secretions, saliva in secreted form and may become the body’s first line of defense against bacteria and viruses [3]. SPLUNC1 is the decisive element in innate immune defense against pathogenic organisms in the respiratory tract infected pulmonary mucosa [4]. SPLUNC1 plays an important role in the antibacterial and anti-inflammatory response to respiratory tract infection caused by Gram-negative bacteria [3]. Besides that, SPLUNC1 plays a very important role in maintaining the steady-state aspects of upper respiratory tract [5]. However, there is no report about the SPLUNC1 sequence of Xinjiang Wild Argali. To study the difference in nucleotide sequences and amino acid sequences of SPLUNC1 gene between wild and domestic animals, the present study used the sequences from cattle and sheep as the references to clone and analyze the cDNA sequence by RACE technique and in vitro expression in eukaryotic cells, which provides a basis for further studies on the differences in gene functions and disease resistance.
Oral palate mucosa materials were collected by scrapping oral palate from four healthy wild Argali and stored in Trizol (Invitrogen, Beijing, China). Total RNA was extracted by using the Trizol (Invitrogen, China) method and stored at −70°C until use.
The 5′ ends were generated according to the instructions for the 5′ Full RACE Core Set cDNA Kit (Clontech, Beijing, China). Briefly, cDNA was synthesized by reverse transcription (RT) UPM primer (as provided in the kit). Based on the published mRNA sequence of Bos taurus (accession no. NM_174426.3), gene specific primers namely R2 (5′ATTGACCAGNGGGCAC AC3′) and K2(5′CAGGCTGCCAGGGGAGTG3′) were employed in the nested polymerase chain reaction (PCR) at concentration of 20 pmol/μL each. The second amplification products were recovered using ‘Takara Agarose gel DNA purification Kit version 2.0’ according to the manufacturer’s instructions and ligated into pMD-18T cloning vector (Takara Biotechnology, Dalian, China) as described [6]. The sequences in the clones were verified by the sanger DNA sequencing method using Taq DNA Polymerase (Qiagen, Shanghai, China).
The 3′ends were synthesized according to the instructions in 3′Full RACE Core Set cDNA Kit (Takara Biotechnology, China). The 3-sites adapter Primer (provided in the Kit), the P1 primer (5′CATCGTCTCTATGTCACC3′) and P2 primer (5′ATTGAC CAGNGGGCACAC3′) were used at concentration of 20 pmol/μL each. The 3′ end PCR products were purified and ligated into pMD-18T cloning vector (Takara Biotechnology, China). The sequences in the clones were verified by the Sanger DNA sequencing method using Taq DNA Polymerase (Qiagen, China).
The complete coding sequence was generated by RT-PCR as described [6]. The primers used were as follows: SPB1(5′TACGTAATGCACCACCACCACCACCACCTGCTAGAAGCCCT GCCCG3′); SPB2(5′GCGGCCGCTCAGACTTTGATGACAAATTCTAGCCC3′) at concentrations of 20 pmol/μL each. The purified PCR products were ligated into pMD-18T cloning vector (Takara Biotechnology, China). The sequences in the clones were verified by the Sanger DNA sequencing method using Taq DNA Polymerase (Qiagen, China).
The expression vector of recombinant SPLUNC1 protein was constructed by inserting full length SPLUNC1 open reading frame (ORF) into pPIC9K plasmid (Invitrogen, China). Briefly, recombinant pMD-18T with SPLUNC1 ORF was digested with SnaB I and Not I and the SPLUNC1 cDNA fragment was inserted into pPIC9K vector previously linearized with similar enzymes to obtain the pPIC9K/SPLUNC1 cDNA expression vector. The recombinant vector was transformed into Escherichia coli and eight positive clones were verified by enzyme digestion. The recombinant pPIC9K/SPLUNC1 vector was linearized by using Sac I and transformed into Pichia pastoris GS115 by using electroporation-mediated method. His+ transformants were selected on minimal dextrose (MD) plates and selected subsequently on yeast extract peptone dextrose (YPD) plates containing gentamycin 418 (G418; 0.25 to 3 mg/mL), and the genomic DNA of the His+ and G418+ transformants were extracted and analyzed by PCR using the 5′AOX1 primer and the 3′AOX1 primer.
A monoclonal strain that was picked from plate G418 was inoculated into BMGY medium [BMGY: 20 g tryptone, 10 g yeast extract, 100 mL 10×yeast nitrogen base (YNB W/O amino acids), 2 mL 500×D-Biotin (B), 100 mL 10× glycerol (Gl), 100 mL 1 M phosphate buffer, 700 mL H2O], and cultured for 2 days (OD600 = 2–6) in the shaker. Pichia pastoris liquid was centrifuged. Then, it was transferred into BMMY medium [BMMY: 20 g tryptone, 10 g yeast extract, 100 mL 10×YNB, 2 mL 500×B, 100 mL 10× methanol (M), 100 mL 1 M phosphate buffer, 700 mL H2O], which was used for inducing protein expression. Methanol was added to a final concentration of 0.5% (v/v) every 24 h. After 3 days, yeast liquid was collected and centrifuged. Finally, the supernatant was collected separately and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Following induction, the supernatant were collected were isolated as described by the manufacturer (Qiagen, China). Briefly, yeast liquid which contained the expression product was collected by centrifugation and the supernatant collected. The supernatant was filtered through a 0.22 μm membrane. The filtrate was passed through a Ni-NTA agarose affinity chromatography column with 10 to 15 volumes of binding buffer (pH 8.0 50 mM phosphate buffer containing 0.5 M NaCl and 20 mM imidazole) until the optical density (OD) value reached the 280 nm absorbance, and was collected respectively. Then, it was eluted with elution buffer (pH 8.0 50 mM phosphate buffer containing 0.5 M NaCl and 300 mM imidazole). Finally, the liquid effluent was collected separately and analyzed by SDS- PAGE [7].
After the process of SDS-PAGE electrophoresis, the proteins were electroblotted to polyvinylidene fluoride membrane. Then the membrane was placed in blocking solution about 1 h; incubated at 4°C overnight with Murine monoclonal antibody with anti-His tag (1:2,000), next, the membrane was washed 3 times with Tris-buffered saline and Tween 20 (TBST) (TBST: 50 mL 1 M pH 7.5 Tris HCl, 8 g NaCl, 0.2 g KCl, 0.5 mL Tween 20, 1,000 mL H2O). Incubated at 25°C with the secondary antibody, horseradish peroxidase labeled goat anti-mouse IgG (1:5,000), for about 90 min, then the membrane was washed three times with TBST, and finally colored with diaminobenzidine coloring solution.
Determination of the activity of the expression product of SPLUNC1 was tested by incubating MO, 105 colony-forming units (CFUs)/mL, with recombinant SPLUNC1 (make the final concentration of recombinant SPLUNC1 protein up to 1 μg/mL, 3 μg/mL, 10 μg/mL in every well), phosphate-buffered saline (PBS) (as control) in a 24-well plate for 48 hours at 37°C as described [8,9]. MO RNA was extracted, and real-time PCR was performed to quantify MO 16S rRNA levels. Finally, statistical analysis was carried out for the data by using one-way analysis of variance by SPSS17.0 (www.xpgod.com).
Nucleotide sequencing combined with a search of the GenBank database revealed eight clones generated by 5′RACE-PCR (≈635) (Figure 1A), having no initiator element in their open reading frames. The 3′RACE-PCR products (Figure 1B) had complete 3′ends containing polyadenylation tail of 12A’s at 224 bp downstream of the stop codon TAA. All the transformation clones had inserts of the expected SPLUNC1 cDNA-ORF as identified by SnaB I and Not I enzyme digestion (Figure 2A). The sequences were confirmed in all the nine clones. The 3′, 5′RACE and full length sequences were 635 bp (3′ends), 620 bp (5′ends), and 1,076 bp (SPLUNC1 cDNA) respectively, each having 99% identity to Ovis.aries mRNA.
The sequence of Wild Argali SPLUNC1 cDNA and the inferred amino acids was shown in Figure 2B. The accuracy of the sequence was determined on both strands and found to be in order. The cDNA insert was 1,076 bp long. SPLUNC1 cDNA-ORF contains one long open reading frame of 768 bp from 64 nt to 831 nt. The first methionine codon ATG begins at 64 to 66 nt and ends at presumed stop codon TAA at 829 to 831 nt. The bases AATAAA at 1,034 to 1,039 nt downstream are the distal polyadenylation stop signals for SPLUNC1 cDNA. However, the TATA box was not found in the upstream of SPLUNC1 cDNA-ORF.
The initial ATG and stop TAG codons are shaded. The distal polyadenylation signal sequence AATAAA are shaded. The nucleotide and its amino acids are numbered along the left margins respectively.
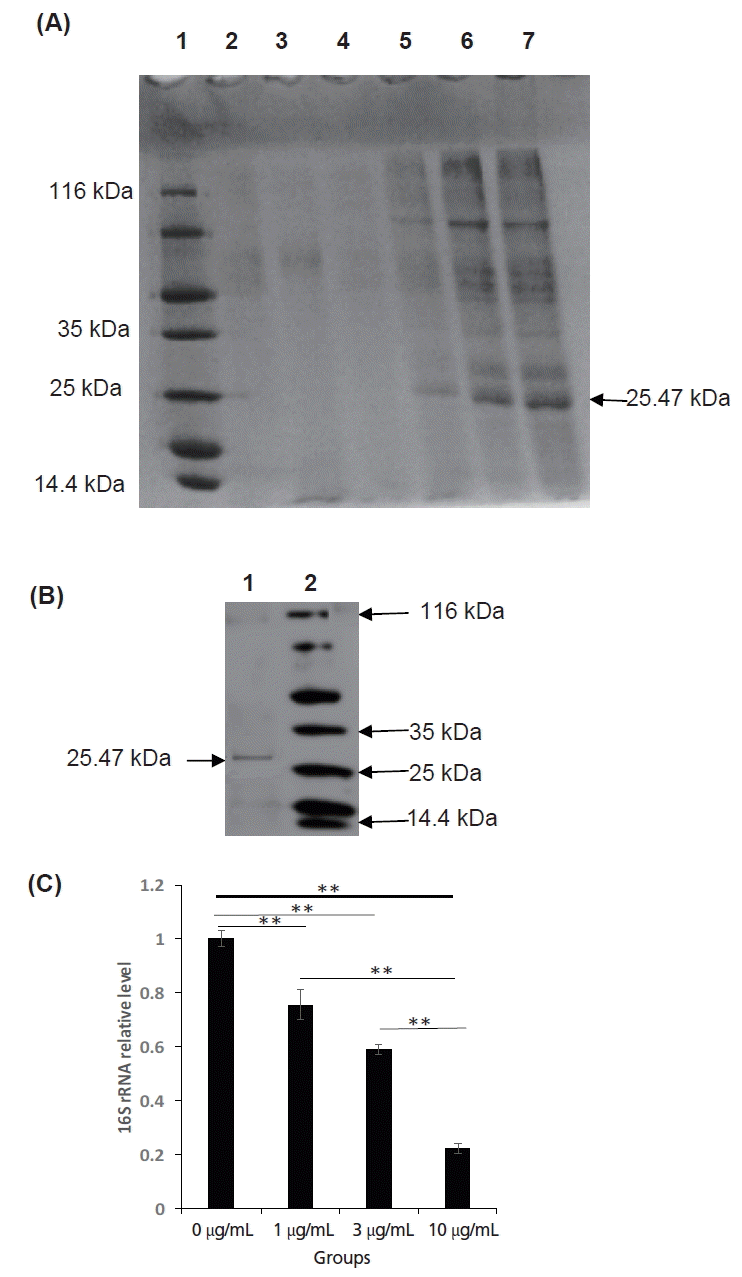
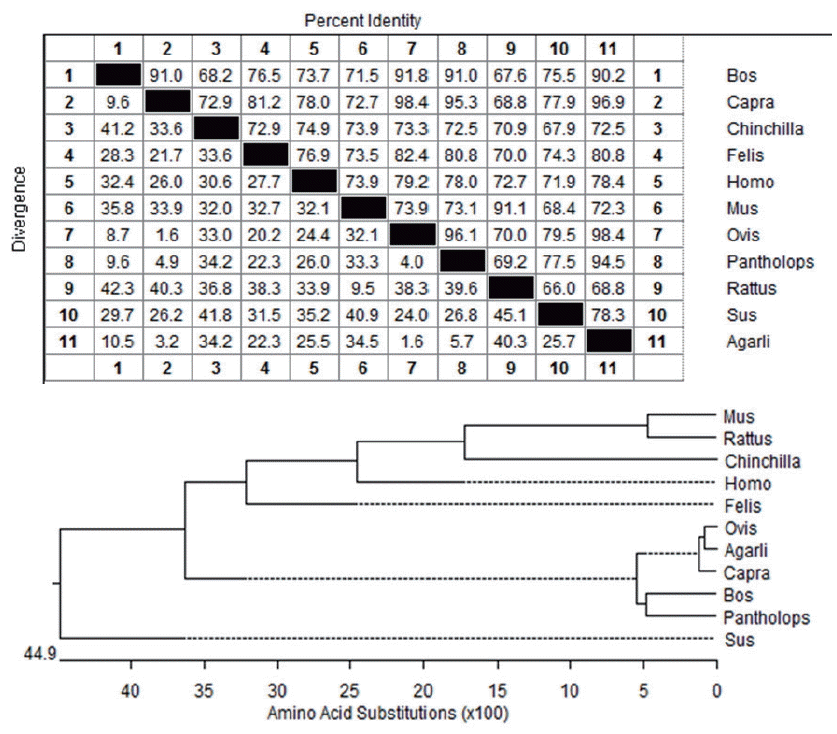
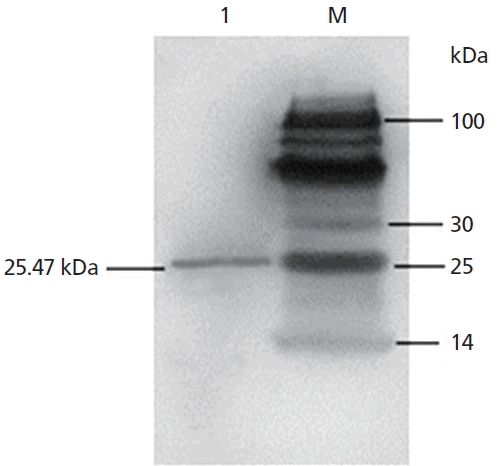
The open reading frame encoded a 255 amino acid residue protein with an isoelectric point (pI) of 5.07 and a predicted molecular mass of 26,486.43 Da (≈26.49 kDa). The recombinant protein corresponded to the expected molecular mass of 25.47 kDa as judged by SDS-PAGE and the expression product was detected in the supernatant of the Pichia pastoris (Figures 3A, B). The protein consists of 118 hydrophobic amino acids (Ala, Ile, Leu, Phe, Trp, and Val), 57 Polar amino acids (Asn, Cys, Gln, Ser, Thr, and Tyr), 12 strongly basic amino acids (Lys and Arg) and 19 strongly acidic amino acids (aspartic acid [Asp] and glutamic acid [Glu]). Alignment of amino acid sequences of SPLUNC1 cDNA against amino acid sequences from other species revealed SPLUNC1 protein shared 98.4%, 96.9%, 94.5%, 90.2%, 80.8%, 78.4%, 78.3%, 72.5%, 72.3%, 68.8% identity with those of SPLUNC1 cDNA from Ovis aries (accession no. NP_ 001288334.1), Capra hircus (accession no. XP_005688516.1), Pantholops hodgsonii (accession no. XP_005979709.1), Bos taurus (accession no. NP_776851.1), Felis catus (accession no. XP_0069 29910.1), Homo sapiens (accession no. NP_001230122.1), Sus scrofa (accession no. NP_001005727.1), Chinchilla lanigera (accession no. NP_001269294.1), Mus musculus (accession no. NP_035256.2), and Rattus norvegicus (accession no. NP_742028.1), respectively (Figure 4). The phylogenetic tree generated from nucleotide sequences (Figure 4) revealed a close relationship between Argali and Ovis aries.
The results showed that a distinct protein bands appeared at 25.47 KDa place (Figure 5) and the recombinant proteins can bind specifically His tag antibody by Western blot analysis, so the expression of recombinant SPLUNC1 protein was confirmed.
The results from the test of recombination SPLUNC1 protein on MO showed that the expression product could inhibit MO proliferation very well (p<0.01), which proved the expressed protein had biological activity (Figure 3C).
The SPLUNC1 cDNA sequence of the Xinjiang Wild Argali for test was obtained by RT-PCR, 5′RACE, and 3′RACE methods. The SPLUNC1 cDNA was 1,076 bp long with an open reading frame of 768 bp, which encoded a 26.49 kDa protein composed of 255 amino acids, containing one initiation codon ATG and one termination codon TAA. The 5′UTR ends at 63 nt and the 3′UTR ends at 245 bp in Argali, while the 5′UTR ends at 58 nt, the 3′UTR ends at 265 bp in sheep. The amino acid sequence homology of Argali and sheep is 98.4% by basic local alignment search tool (BLAST). When we designed primer of expression of Argali SPLUNC1 protein, removed the signal peptide, and added His tag sequence, thus the target protein’s molecular weight becomes 25.47 kDa.
Bacterial infection in the lung is a major cause of mortality and morbidity, especially in high-risk groups such as immunocompromised mammalian species, the young and those with other underlying pulmonary diseases such as pneumonias. The antimicrobial function of SPLUNC1 probably plays a critical role in host defense against pathogens not only in maintaining homoeostasis of healthy individuals but also in protecting mammal whose lungs are compromised by a chronic lung disease. Results from both in vitro and in vivo studies indicate the antimicrobial function of SPLUNC1 and suggest a defensive role of SPLUNC1 in airways exposed to bacterial infection [10].
SPLUNC1 has been shown to be a key role in promoting host innate immunity against Mycoplasma pneumoniae (Mp), and previous studies have verified in mouse. Recombinant mSPLUNC1 protein inhibits Mp growth in a dose-dependent manner [11].
Using SPLUNC1 knockout and transgenic mice to test this model against Mp in vivo, found that SPLUNC1 was highly expressed in the trachea and intrapulmonary, and undetectable in any tissues or bronchoalveolar lavage (BAL) fluid harvested from SPLUNC1−/− mice, Mp levels in SPLUNC1−/− mouse lungs were threefold higher than those in the SPLUNC1+/+ littermate control mice after Mp infection 24 h. Meanwhile, hSPLUNC1 overexpression in mouse airways resulted in doubling of SPLUNC1 protein in BAL fluid, lung Mp load in hSPLUNC1 transgenic mice was about threefold lower than that in the wild-type littermate control mice after Mp infection. A critical in vivo role of SPLUNC1 was demonstrated in host defense against Mp [12]. So we tested the activity of Argali recombinant SPLUNC1 protein by using real-time PCR in this research, found that Argali recombinant SPLUNC1 protein could remarkably inhibit the growth of MO, have biological activity, and lay the foundation of subsequent function study for this gene.
Mycoplasma pneumonia infections (also known as infections of pleuropneunonia of sheep and goats), have clinical features of a high fever, cough, chest and pleural serous and fibrinous inflammation, can be acute and chronic, and has a high mortality. The main method for the treatment of the disease is with antibiotics [5,13]. There is a difference in sensitivity of MO to antimicrobial agents, resulting in a persistent disease in the sheep after the drug was administered. However, Argali recombinant SPLUNC1 protein could remarkably inhibit the growth of MO (data not shown), so there are good application prospects for development and utilization of Argali SPLUNC1 gene and treatment of infectious diseases of lungs in sheep.
ACKNOWLEDGMENTS
The research work was funded by a grant from the National Natural Science Foundation of P. R. China (NO: 31460686).
Figure 1
The 5′-RACE and 3′-RACE PCR products of SPLUNC1. (A) The 5′-RACE-PCR products, Key: Lane 1: Positive 3′ends PCR products ≈635 bp; Lane 2: DNA Maker I molecular marker. (B) The 3′-RACE-PCR products, Key: Lane 1: positive 5′ends PCR product ≈620 bp; Lane 2: DNA Maker I molecular marker. PCR, polymerase chain reaction.

Figure 2
Restriction enzyme analysis of expression vector of pPIC9K-SPLUNC1 and the sequence of SPLUNC1 cDNA. (A) The recombinant plasmids of eight clones containing inserts of the full-length SPLUNC1 cDNA-ORF identified by SnaB I and Not I enzyme digestion. Key: Lane 1: ≈744 bp SPLUNC1-ORF; Lane 2: empty vector DNA; Lane 3: DNA Maker I molecular marker. (B) Nucleotide and predicted amino acid sequence of SPLUNC1cDNA. The complete SPLUNC1cDNA sequence consisting of 1,076 nucleotides and open reading frame of 768 bp. The initiation ATG and stop TAA codons are shaded. The distal polyadenylation signal sequence AATAAA are shaded. The nucleotide and its amino acids are numbered along the left margins respectively. The putative signal peptide sequence is shown as capital italicized letters.
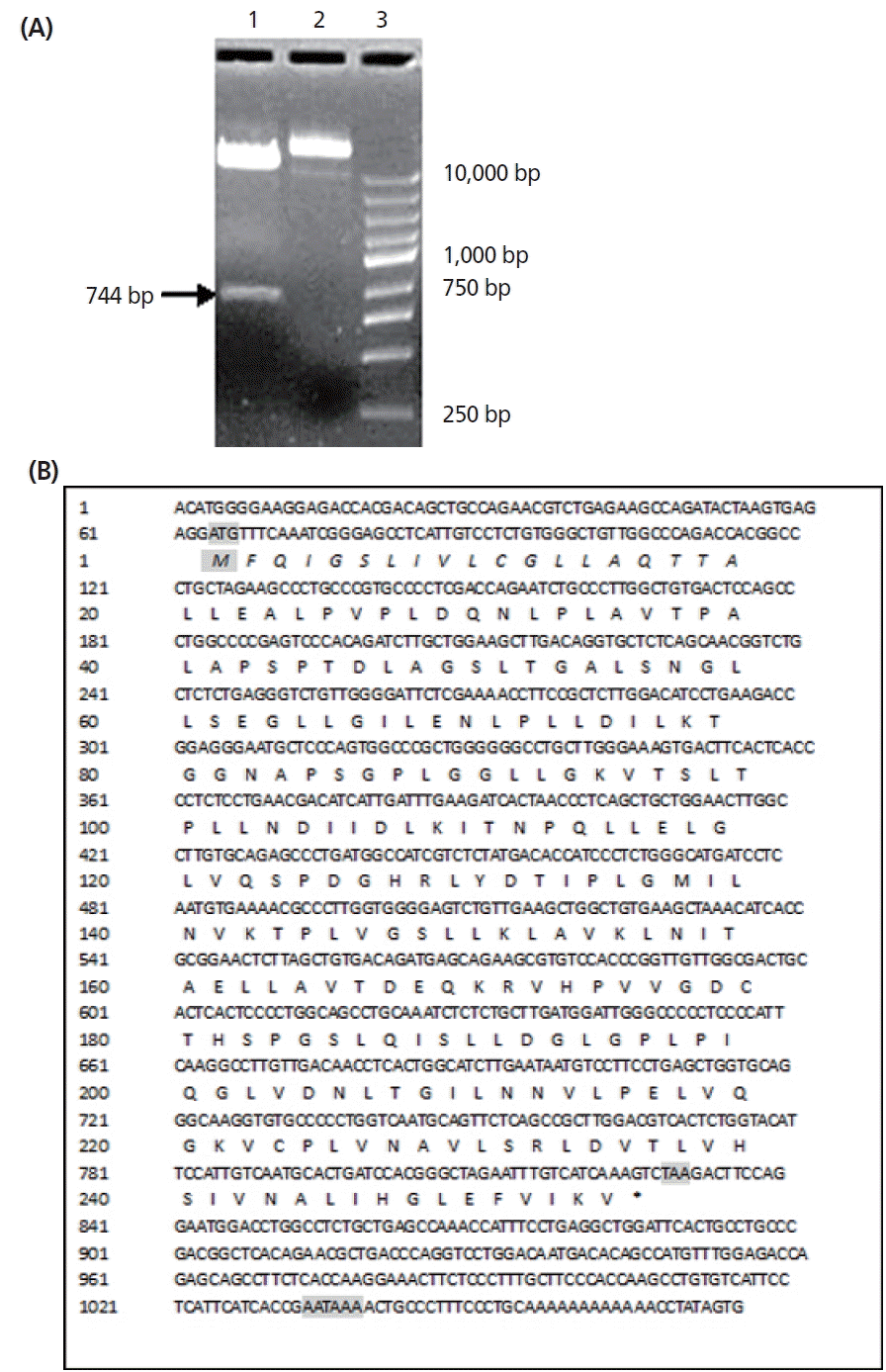
Figure 3
Expression, purification and biological activity analysis of SPLUNC1. (A) Expression of SPLUNC1 protein. Gels were stained with Coomassie blue and 20 μL samples were loaded perlane. Key: Lane 1: standard protein molecular weight marker; Lane 2: pPIC9K empty expression vector (negative control); Lane 3–7: transformed Pichia pastoris supernatant after 0, 24, 48, 72, 96 h of induction with methyl alcohol, respectively. (B) Purification of SPLUNC1 protein. Gels were stained with Coomassie blue and 10 μL samples were loaded per lane. Key: Lane 1: purified recombinant 25.47 kDa SPLUNC1 protein; Lanes 2: tandard protein molecular weight marker. (C) The effect analysis of Mycoplasma ovipneumoniae growth in recombinant SPLUNC1 protein of Argali.
REFERENCES
1. Yu YQ, Ji MZ, Liu CG, Li KC, Guo SG. Geographical distribution and vicissitude of argali, Ovis ammon, in China. Biodivers Sci 2008; 16:197–204.

2. Sun YM, Chen KL, Shen W, Cui RP, Lu HF. Cloning and sequence analysis of wild argali ISG15 cDNA. Asian-Australas J Anim Sci 2014; 27:561–6.



3. Di YP, Harper R, Zhao Y, et al. Molecular cloning and characterization of spurt, a human novel gene that is retinoic acid-inducible and encodes a secretory protein specific in upper respiratory tracts. J Biol Chem 2003; 278:1165–73.


4. Liu Y, Bartlett JA, Di ME, et al. SPLUNC1/BPIFA1 contributes to pulmonary host defense against Klebsiella pneumoniae respiratory infection. Am J Pathol 2013; 182:1519–31.



5. McGillivary G, Bakaletz LO. The Multifunctional host defense peptide SPLUNC1 is critical for homeostasis of the mammalian upper airway. PloS ONE 2010; 5:e13224



6. Sambrook J, Russell WD. Molecular cloning: A laboratory manual. 3rd editionCold Spring Harbor: NY: Cold Spring Harbor Laboratory Press; 2001.
7. Marion MB. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248–54.


8. Chu HW, Gally F, Thaikoottathil J, et al. SPLUNC1 regulation in airway epithelial cells: role of toll-like receptor 2 signaling. Respir Res 2010; 5:155–62.



9. Xiao DL, Zhong LL, Liang M, et al. Expression of SPLUNC1 in human bronchial epithelial cells induced by Mycoplasma peumoniae lipid-associated membrane proteins in human bronchial epithelial cells. J Clin Res 2013; 30:1041–5.
10. Di YP. Functional roles of SPLUNC1 in the innate immune response against Gram-negative bacteria. Biochem Soc Trans 2011; 39:1051–5.



11. Chu HW, Thaikoottathil J, Rino JG, et al. Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J Immunol 2007; 179:3995–4002.


12. Gally F, Di YP, Smith SK, et al. SPLUNC1 promotes lung innate defense against Mycoplasma pneumoniae infection in mice. Am J Pathol 2011; 178:2159–67.



13. Hao RX, Hao YQ, Xu CG, Liu B, Zhang C. Isolation of Mycoplasma ovipneumoniae a sheep identification and culture characteristics. J Inner Mongolia Agric Univ 2013; 34:10–3.
- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 6,535 View
- 90 Download
- Related articles
-
Cloning and Sequence Analysis of Wild Argali ISG15 cDNA2014 April;27(4)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




