 |
 |
| Anim Biosci > Volume 30(5); 2017 > Article |
|
Abstract
Objective
This study was performed to reveal the molecular structure and expression patterns of horse glutamate-cysteine ligase catalytic subunit (GCLC) and glutamate-cysteine ligase modifier subunit (GCLM) genes whose products form glutamate cysteine ligase, which were identified as differentially expressed genes in the previous study.
Methods
We performed bioinformatics analyses, and gene expression assay with quantitative polymerase chain reaction (qPCR) for horse GCLC and GCLM genes in muscle and blood leukocytes of Thoroughbred horses
Results
Expression of GCLC showed the same pattern in both blood and muscle tissues after exercise. Expression of GCLC increased in the muscle and blood of Thoroughbreds, suggesting a tissue-specific regulatory mechanism for the expression of GCLC. In addition, expression of the GCLM gene increased after exercise in both the blood and muscle of Thoroughbreds.
The aerobic respiration of muscle tissue is an important process in mammals. In steady-state muscle tissue, as part of the ATP generation mechanism during aerobic respiration, cytochrome c, an essential component of the electron transport system, stabilizes cells that are reacting with oxygen [1]. During rapid exercise, an oversupply of oxygen in the blood reacts with electrons that are generated from the electron transport system and forms reactive oxygen species (ROS) [2]. These ROS, which indicate oxidative stress, destroy DNA through muscle fatigue, damage, and apoptosis [3]. The protection of cellular organelles from ROS occurs in part via the oxidation of glutathione by ROS. The glutamate-cysteine ligase modifier subunit (GCLM) and the glutamate-cysteine ligase catalytic subunit (GCLC) form the glutathione cysteine ligase (GCL) that catalyzes the conversion of glutamate and cysteine into γ-glutamyl cysteine, a glutathione precursor [4]. During strenuous exercise and in other situations that induce oxidative stress, the GCLC gene is expressed [5,6]. In experiments using blood from marathon runners, glutathione expression was found to increase after strenuous exercise, and it has been hypothesized that GCLC was up-regulated in response to this exercise [7]. In the case of GCLM, the expression pattern also increased during strenuous exercise [6], again owing to oxidative stress [8]. A previous study showed that the expression of the GCLC and GCLM genes was up-regulated in the muscle of Thoroughbred horses in response to exercise [9]. These data indicate that glutathione-synthesis-related genes play important roles in alleviating the stress response during exercise. Despite various experiments with GCLC and GCLM, understanding these genes in horses during exercise is still insufficient.
In this study, we performed bioinformatic analyses of horse GCLC and GCLM genes, and investigated their expression in muscle and blood in response to exercise stress to gain insights into their potential application as biomarkers for ROS responses in horses.
Two stallions and one mare Throughbred horses aged 5, 9, and 10, weighing from 500 to 700 kg which were maintained at Ham-an Racing Horse Resort and Training Center were used to obtain the blood and skeletal muscle samples before and after exercise. Exercise was performed by trotting at the speed of 13 km/h for 30 min and cantering through lunging and long-reining (circular bridge lunging).
Three Jeju horses (3 mares), which were maintained at The National Institute of Subtropical Agriculture, Rural Development Administration were used to obtain tissue samples skeletal muscle, kidney, thyroid, lung, appendix, colon, spinal cord and heart. All tissue samples were kept in a liquid nitrogen tank until extraction of RNA was done. All procedures were conducted by following the protocol that had been reviewed and approved by the Institutional Animal Care and Use Committee at Pusan National University (protocol numbers: PNU-2013-0417, PNU-2013-0411).
Total RNA samples for investigation of GCLC and GCLM gene expression were collected from three Thoroughbreds. Skeletal muscle, kidney, heart, liver, lung, colon, and spinal cord tissues were extracted for polymerase chain reaction (PCR) analysis. The various tissues sampled from the horses (50 to 100 mg, or 3 mL in the case of blood) were crushed with a mortar and pestle and mixed with 9 mL of red blood cell lysis buffer (Solgent Co. Ltd., Daejeon, Korea) to remove red blood cells. The cells were then dissolved using 1 mL of TRIzol (Invitrogen, Karlsruhe, Germany), and 200 μL of chloroform was added to remove cells from the organic solvent. The mixture was then shaken for 10 s and left at 4°C for 5 min. Centrifugal separation was carried out at 4°C for 15 min, and then the supernatant was removed to a new test tube and mixed with the same amount of isopropanol. The test tube was left at 4°C for 15 min to produce RNA pellets. Isopropanol was removed by carrying out centrifugal separation at 4°C for 15 min and the sample was then sterilized with 85% ethanol and dissolved in RNase-free water. The purity of the extracted RNA was confirmed by measuring the absorbance at 230 nm and 260 nm using a spectrophotometer (ND-100, Nano Drop Technologies Inc., Wilmington, DE, USA), and only the extracted RNA with purity optic density (OD) value at 230 nm divided by OD value at 260 nm) over 1.8 (found via quantitative analysis) was used. The selected RNA was stored at −70°C until the experiment was performed.
In order to synthesize cDNA, 2 μg of RNA, 1 μL of oligo-dT (Invitrogen, Germany), and 1 μL of RNase-free water were added, the mixture was denatured at 80°C for 3 min, and then the cDNA was synthesized using 4 μL of 5× reverse transcription buffer, 5 μL of 2 mM deoxynucleotides (dNTPs), 0.5 μL of RNase inhibitor (Promega Corporation, Madison, WI, USA), and 1 μL of moloney murine leukemia virus reverse transcriptase (Promega, USA).
The nucleotide sequences of horse GCLC and GCLM genes from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and the Ensembl Genome Browser (http://www.ensembl.org) were retrieved to design the primers with PRIMER3 software (http://bioinfo.ut.ee/primer3-0.4.0/). The primers sequences of horse GCLC and GCLM genes were as followed: GCLC forward 5′-ACCAGGGTGATCCTGTCGTA-3′ and reverse 5′-ATCCCGTTTCTGTGCGACTT-3′, and GCLM forward 5′-GTG TGA TGC CAC CTG ATT TG-3′ and reverse 5′-GCT TTT CAC GAT GAC CGA GT-3′. The PCR to amplify the target genes on the cDNA was carried out under the following conditions: 1.8 μL dNTP, 2 μL 10× buffer, 0.2 μL HS-Taq, and 12 μL distilled water were added to 2 μL, 50 ng/μL diluted DNA and 5 pmol/μL diluted forward primer and reverse primer. The PCR was carried out in a total volume of 20 μL. The PCR procedure was: denaturation at 94°C for 10 min, and a second denaturation at 94°C for 30 s, followed by annealing at 58°C for 30 s, and extension at 72°C for 30 s. This was repeated for 40 cycles, and then a final extension was performed at 72°C for 10 min. The band was confirmed on UV using a 1.5% SeaKem LE agarose gel (Lonza, Rockland, MD, USA).
The real time quantitative polymerase chain reaction (RT-qPCR) was performed using a C1000 Thermal Cycler (Bio Rad, Hercules, CA, USA) in order to measure the relevant expression of the target genes. A reaction solution of 25 μL was used and the solution was prepared as follows: 2 μL and 5 μL of distilled water, and 2 μL of diluted cDNA (50 ng/μL) were added to 14 μL of SYBR green master mix (Bio Rad, USA), along with 5 pmol/μL each of diluted forward primer and reverse primer. The RT-qPCR conditions were as follows: the first denaturation was carried out at 94°C for 10 min, and then the second denaturation was at 94°C for 10 s; the annealing occurred at 60°C for 10 s, followed by an extension at 72°C for 30 s. This was carried out repeatedly 40 times. All measurements were performed in triplicate for all specimens, and the comparative method used was the 2−ΔΔCt method [10]. The relevant expression of the target genes was calculated using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a normalizer.
The amino acid sequences of GCLC and GCLM of various species were obtained from National Center for Biotechnology Information (NCBI): Amino sequence of GCLC was obtained with cow (NP_001077143.1), human (NP_001489.1), macaque (XP_ 005552782.1), mouse (NP_034425.1), orangutan (XP_002817060.1), rat (NP_036947.1), pig (XP_003482212.1), horse (XP_001503230.2) and cow (NP_001033232.1), human (NP_002052.1), macaque (NP_001270732.1), mouse (NP_032155.1), orangutan (XP_ 002810638.1), rat (NP_059001.1), pig (XP_001926413.1), horse (F6WKI2) was obtained for GCLM. Amino acids were aligned using Multiple Sequence Comparison by Log-Expectation (MUSCLE) (http://www.ebi.ac.uk/Tools/msa/muscle/). Phylogenetic analysis was performed using Neighbor-Joining method [11] with pairwise deletion, 1,000 bootstrap replication, and Kimura 2 as described previously [12].
As human and mouse, horse GCLC and GCLM are encoded by different genes which are located in chromosome 20 and 16 respectively. The genomic structure of horse GCLC and GCLM is shown in (Figure 1). The GCLC gene consists of 16 exons, and the GCLM gene is composed of 6 exons. The full lengths of the GCLC and GCLM genes are 1,935 and 1,425 bp, respectively. The horse GCLC gene encodes 637 amino acids and the GCLM gene encodes 233 amino acids (Table 1).
To investigate the evolutionary relationships of GCLC and GCLM in horses, we obtained gene sequences from eight species of mammals (human, macaque, orangutan, cow, pig, horse, rat, and mouse) from Ensembl 62 and conducted a phylogenetic analysis (Figure 2). The amino acid similarity of the two genes in the eight mammals is over 90% (Tables 2, 3). Both horse GCLC and GCLM are clustered in the same clade as other livestock such as pig and cow. Simple modular architecture research tool (SMART) analysis revealed that horse GCLC protein contains the gamma-glutamylcysteine synthetase (GCS) domain from amino acid position 236 to 608, which represents a signature catalytic subunit of glutamate-cysteine ligase domain (EC 6.3.2.2) (Figure 1B). Horse GCLM protein possess the aldo-keto reductase family domain which is covered by the amino acid sequences from 29 to 190 (Figure 1B). Taken together, these two proteins are structurally conserved, suggesting that the function of GCLC and GCLM are conserved during the evolution.
To get the insights for function of these genes, we conducted RT-PCR and RT-qPCR using seven Thoroughbred horse tissues (Figure 3). The results showed that GCLC and GCLM are expressed in skeletal muscle, kidney, heart, liver, lung, colon, and spinal cord, suggesting ubiquitous expression. But there were no differences in both of GCLC gene and GCLM gene expression in the horse tissues studied in this study (Figure 3). To reveal the relationship between exercise induced stress and transcriptional activity of glutathione synthesis related genes, we prepared total RNAs from skeletal muscle and blood of Thoroughbred horses and performed RT-qPCR analysis for GCLC and GCLM genes. As result, it was found that the expression of both of GCLM and GCLC genes was up-regulated in skeletal muscle as well as blood leukocytes.
The mRNA level of GCLC gene after exercise was about 10-fold higher than that before exercise in skeletal muscle (Figure 4A left; p<0.05), whereas the degree of increase of mRNA level in blood leukocytes is less compared to skeletal muscle (Figure 4B right; p<0.001). The level of mRNA of horse GCLM gene also increased in both skeletal muscle and blood leukocytes in response to exercise (Figure 4B). In either case, it is not clear whether the increased expression of mRNA of these genes is the consequences of increased transcription efficiency or post-transcriptional modifications, i.e., increased stability, and is also not certain whether increased mRNA level will result in increased protein level, leading to augmented GCL activity. In a previous study, GCLC contents and activity tended to increase with exercise in skeletal muscle of both old and adult rats with different kinetics, indicating differential regulation of GCLC content and activity. Nonetheless, in old rats, GCLM contents seemed to increase more but not in adult rats [13,14]. As the expression of GCLM and the association of GCLM with GCLC are responsible for the increase GCL activity [15] and ROS rapidly activates GCL activity by facilitating the holozyme formation [5], it is reasonable to assume that GCL might be activated in skeletal muscle and blood leukocytes of Thoroughbred horses in response to exercise stress to alleviate stress induced ROS.
In this study, we observed that amino acid sequences of GCLC and GCLM are highly conserved evolutionary and, especially, functional domains of GCS and aldo-keto reductase family domain are well conserved in horse proteins, suggesting functional conservation of these proteins. It is of note that the mRNA level of GCLC and GCLM gene is increased in both skeletal muscle and blood leukocytes in response to exercise. These results will provide the basic foundation for the future studies to reveal the importance of GCL activity to relieve ROS stress caused by exercise in racing horse, leading better interventions to racing stresses and the development of reliable non-invasive biomarkers for ROS responses.
ACKNOWLEDGMENTS
This work was supported by a grant from the Next-Generation BioGreen 21 Program (No.PJ01104401, PJ01117301), Rural Development Administration, Republic of Korea, and research funds for newly appointed professors of Chonbuk National University in 2016 (K.D. Song).
Figure 1
i) Gene structure of the glutamate-cysteine ligase catalytic subunit (GCLC) and glutamate-cysteine ligase modifier subunit (GCLM) genes in horses. (A) Gene structure of GCLC and GCLM. GCLC has 16 exons, and GCLM has 6 exons. Black boxes indicate exons, grey boxes indicate untranslated regions (UTR), and black lines indicate introns.
ii) Gene structure of the glutamate-cysteine ligase catalytic subunit (GCLC) and glutamate-cysteine ligase modifier subunit (GCLM) genes in horses (Continued). (B) Amino acid sequence alignment of GCLC and GCLM proteins with MUSCLE software (http://www.ebi.ac.uk/Tools/msa/muscle/).
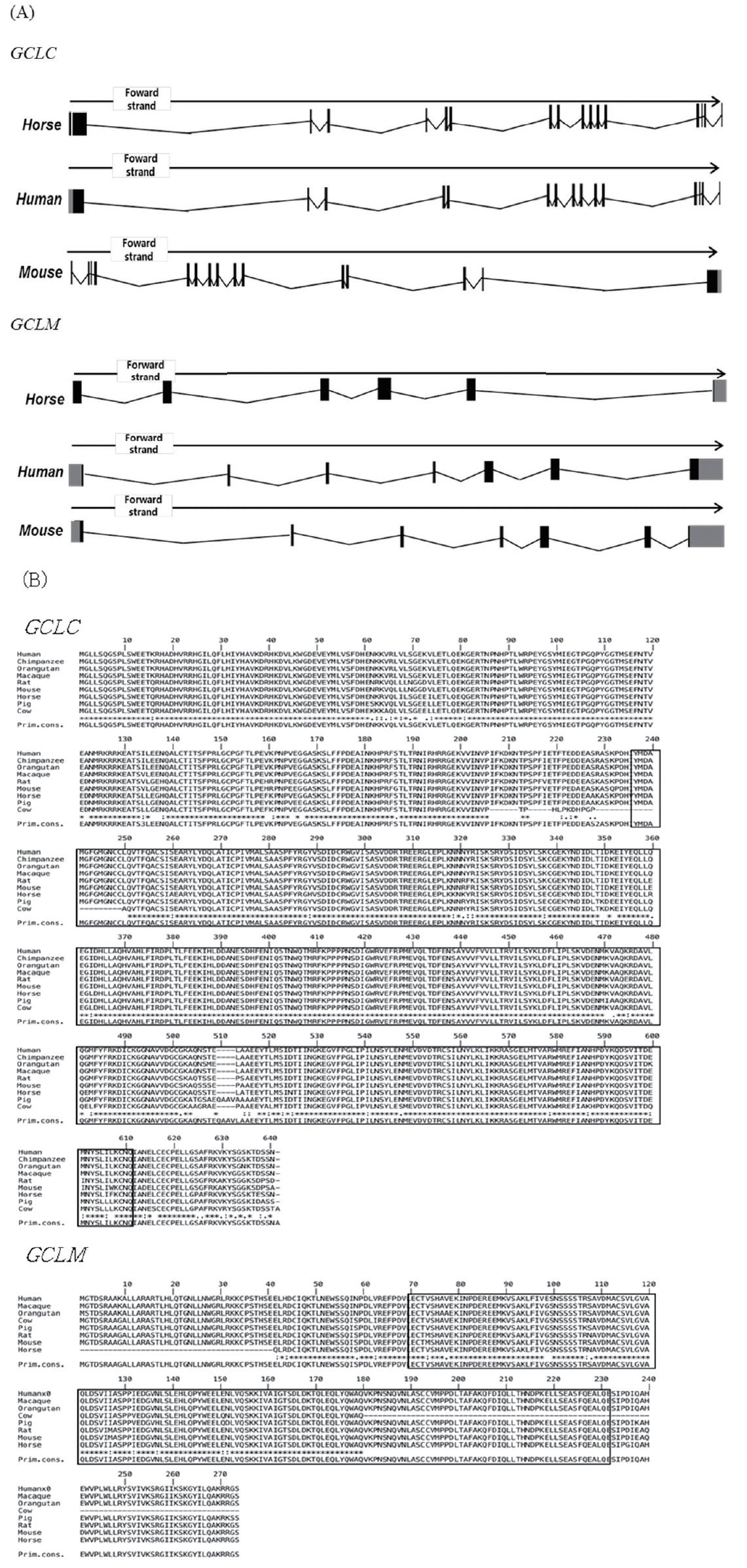
Figure 2
Unrooted phylogenetic trees for horse glutamate-cysteine ligase catalytic subunit (GCLC) genes and glutamate-cysteine ligase modifier subunit (GCLM) genes based on amino acid sequences.
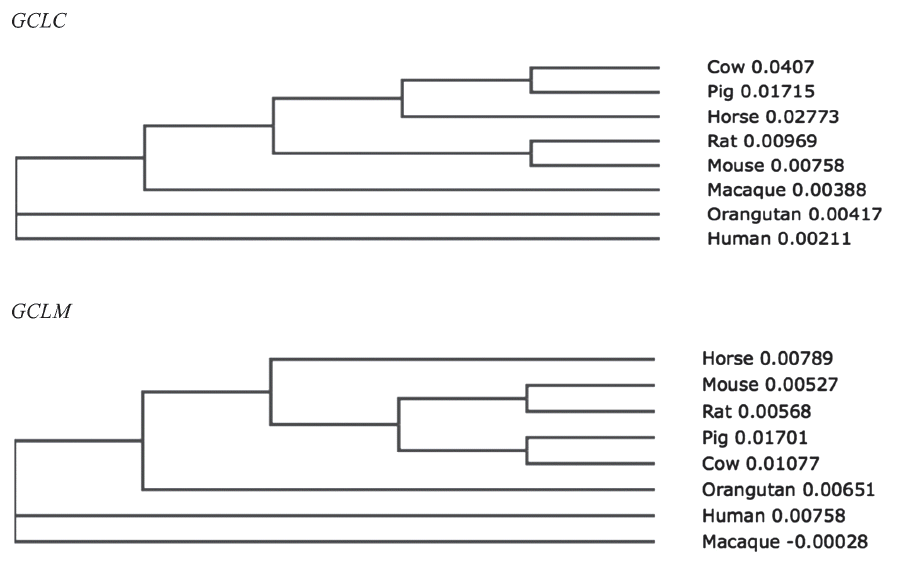
Figure 3
Expression of horse glutamate-cysteine ligase catalytic subunit (GCLC) gene (A), and glutamate-cysteine ligase modifier subunit (GCLM) gene (B) by RT-qPCR and RT-PCR. mRNA expressions were determined by real-time quantitative polymerase chain reaction analysis (RT-qPCR; top) and RT-PCR (bottom) in a variety of tissues from the Thoroughbred horse. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization.
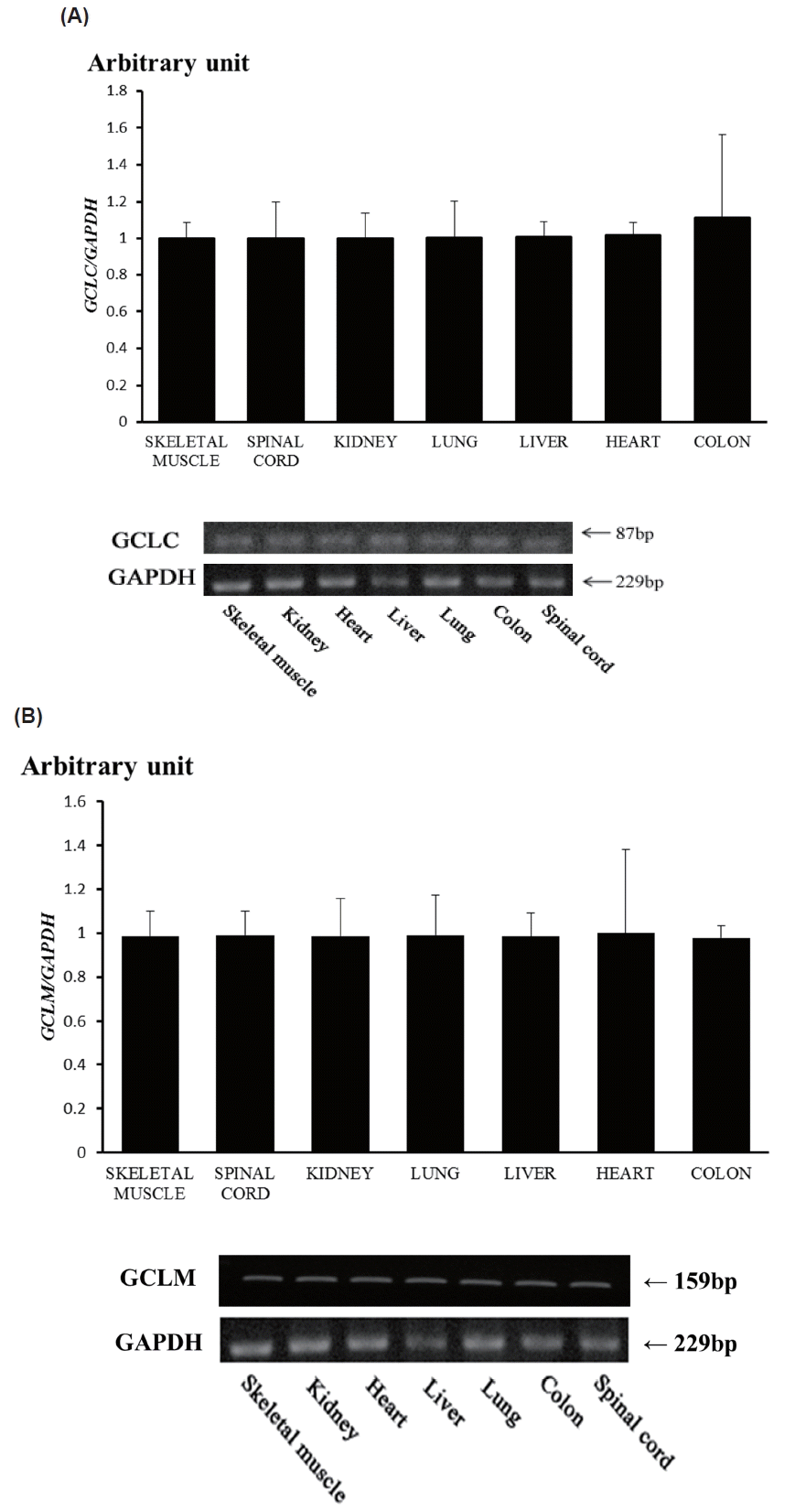
Figure 4
Expression of glutamate-cysteine ligase catalytic subunit (GCLC) and glutamate-cysteine ligase modifier subunit (GCLM) genes determined by real-time quantitative polymerase chain reaction analysis (RT-qPCR) in horses before and 30 minutes after exercise. (A) Expression of the GCLC gene in muscle and blood leukocytes of Thoroughbred horses (n = 3, * p<0.5, *** p<0.001). (B) Expression of the GCLM gene in muscle and blood leukocytes of Thoroughbred horses (** p<0.05,). Quantitative analysis was performed using the 2−ΔΔCt method. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for normalization.
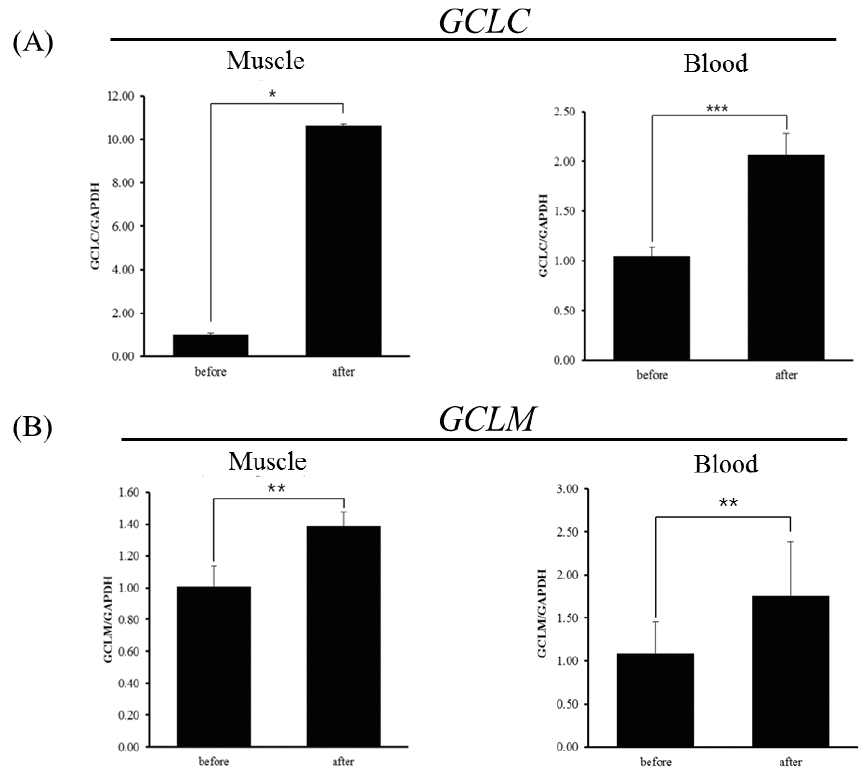
Table 1
Genetic information for the glutamate-cysteine ligase catalytic subunit (GCLC) and glutamate-cysteine ligase modifier subunit (GCLM) genes in horses
| Gene symbol | Location | Length (bp) | Length (aa) | Number of exons |
|---|---|---|---|---|
| GCLC | Chr 20: 50,985,591–51,029,180 | 1,935 | 637 | 16 |
| GCLM | Chr 5: 71,102,814–71,115,917 | 1,425 | 233 | 6 |
Table 2
Amino acid sequence identity of GCLC protein
Table 3
Amino acid sequence identity of GCLM protein
REFERENCES
1. Couto N, Malys N, Gaskell SJ, Barber J. Partition and turnover of glutathione reductase from Saccharomyces cerevisiae: a proteomic approach. J Proteome Res 2013; 12:2885–94.


2. Zhuo Y, Luo H, Zhang K. Leber hereditary optic neuropathy and oxidative stress. Proc Nat Acad Sci USA 2012; 109:19882–3.



3. Powers SK, Kavazis AN, McClung JM. Oxidative stress and disuse muscle atrophy. J Appl Physiol 2007; 102:2389–97.


4. Nava GM, Lee DY, Ospina JH, Cai SY, Gaskins HR. Genomic analyses reveal a conserved glutathione homeostasis pathway in the invertebrate chordate Ciona intestinalis. Physiol Genomics 2009; 39:183–94.



5. Krejsa CM, Franklin CC, White CC, et al. Rapid activation of glutamate cysteine ligase following oxidative stress. J Biol Chem 2010; 285:16116–24.



6. Muthusamy VR, Kannan S, Sadhaasivam K, et al. Acute exercise stress activates Nrf2/ARE signaling and promotes antioxidant mechanisms in the myocardium. Free Rad Biol Med 2012; 52:366–76.



7. Lee KY, Kim SJ, Cha YS, et al. Effect of exercise on hepatic gene expression in an obese mouse model using cDNA microarrays. Obesity 2006; 14:1294–302.


8. Solis WA, Dalton TP, DieterM Z, et al. Glutamate–cysteine ligase modifier subunit: mouse Gclm gene structure and regulation by agents that cause oxidative stress. Biochem Pharmacol 2002; 63:1739–54.


9. Park W, Kim J, Kim HJ, et al. Investigation of de novo unique differentially expressed genes related to evolution in exercise response during domestication in thoroughbred race horses. PloS one 2014; 9:e91418



10. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001; 25:402–8.


11. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406–25.

12. Cho HW, Shin S, Park JW, et al. Molecular characterization and expression analysis of the peroxisome proliferator activated receptor delta (PPARδ) gene before and after exercise in horse. Asian-Australas J Anim Sci 2015; 28:697–702.



13. Chen CNJ, Brown-Borg HM, Rakoczy SG, Thompson LV. Muscle disuse: adaptation of antioxidant systems is age dependent. J Gerontol A Biol Sci Med Sci 2008; 63:461–6.



- TOOLS





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





