 |
 |
Abstract
The average twin lambing rate of Bayanbulak sheep is 2% to 3%. However, a flock of sheep with a close genetic relationship and an average of 2 to 3 lambs per birth has been found recently. To determine the major genes controlling the prolificacy of the flock in the present study, the flock was designated A while 100 normal Bayanbulak sheep were randomly selected to comprise the control flock B. Ligase detection reaction method was applied to detect and analyze the 10 mutational loci of the 3 candidate prolificacy genes including bone morphogenetic protein type I receptors, bone morphogenetic protein 15, and growth differentiation factor 9. The 10 mutational loci are as follows: FecB locus of the BMPR-IB gene; FecXI, FecXB, FecXL, FecXH, FecXG, and FecXR of the BMP15 gene; and G1, G8, and FecTT of the GDF9 gene. Two mutations including BMPR-IB/FecB and GDF9/G1 were found in Bayanbulak sheep. Independence test results of the two flocks demonstrate that the FecB locus has a significant effect on the lambing number of Bayanbulak sheep. However, the mutation frequency of the G1 locus in GDF9 is very low. Independence test results demonstrate that the GDF9 locus does not have a significant impact on the lambing performance of Bayanbulak sheep. Among the 10 detected loci, BMPR-IB/FecB is the major gene that influences the high lambing rate of Bayanbulak sheep.
Fertility is an important trait of animals, specifically for sheep. Three transforming growth members, BMPR-IB, BMP15, and GDF9, are the major genes of high prolificacy. A746G mutation in exon 6 of the bone morphogenetic protein receptor IB gene has been found which induces the 249th amino acid to turn from glutamine (Q) to arginine (R) in Booroola sheep with high prolificacy. The mutation is closely linked to the high prolificacy of Booroola Merino ewes, and the A746G mutation of the BMPR-IB gene located at chromosome 6 has been defined as FecB (Mulsant et al., 2001; Wilson et al., 2001).
By promoting granulosa cell mitosis, bone morphogenetic protein 15 inhibits the expression of the follicle-stimulating hormone receptor in granular cells, and adjusts their proliferation and differentiation. Thus, bone morphogenetic protein 15 plays a key role in the reproductive regulation of mammalian females (Otsuka, 2000; Otsuka, 2001; Juengel, 2002; Moore and Shimasaki, 2005). The BMP15 gene of sheep is located in the X chromosome. Six mutation loci of the BMP15 gene exert a significant effect on the high prolificacy of six sheep breeds, namely, FecXI (Inverdale sheep), FecXB (Belclare and Cambridge sheep), FecXL (Lacaune sheep), FecXH (Hanna sheep), FecXG (Belclare and Cambridge sheep), and FecXR (Rasa Aragonesa sheep) (Galloway, 2000; Hanrahan et al., 2004; Bodin et al., 2007; Martinez-royo et al., 2008; Monteagudo et al., 2009).
Growth differentiation factor 9, a growth factor secreted by the oocyte, has an important regulatory effect on the growth and differentiation of the ovarian follicle. Sheep GDF9 gene is located on chromosome 5. Nine mutations (G1 to G8 and FecTT) on the GDF9 gene are related to prolificacy (Hanrahan et al., 2004; Nicol et al., 2009).
Bayanbulak sheep are a local breed which originated at the Bayangol Mongol Autonomous Prefecture in Xinjiang, China. In addition to good adaptability to the sub-alpine steppe climate with an altitude of 3,000 meters, Bayanbulak sheep possess advantageous attributes such as precocity, crude feed tolerance, and so on. However, the breeding rate of Bayanbulak sheep is 91.5% to 103%, and their twin lambing rate is 2% to 3%. In March 2011, we accidentally found a family (20 head) of Bayanbulak sheep including one progenitor ewe (702), one progenitor ram (707), six daughter ewes and one first-descendant ram of the progenitor ewe (702), five granddaughter ewes, and four daughter ewes and two granddaughter ewes of the progenitor ram (707) (Figure 1). The ewes in this flock produce two or three lambs per lambing. Their multi-lambing rate is 100%. In the current research, the subjects are the normal Bayanbulak sheep flock and the small Bayanbulak sheep flock with high prolificacy. The 10 loci of the 3 genes related to high prolificacy (A746G mutational site on BMPR-IB gene exon 6; FecXI, FecXB, FecXL, FecXH, FecXG, and FecXR mutations on the BMP15 gene; and G1, G8, and FecTT mutations on the GDF9 gene) were detected by ligase detection reaction method (LDR). Single nucleotide polymorphisms of the 10 loci were detected and compared between the normal flock and the high-prolificacy Bayanbulak sheep flock, respectively. The current paper aims to determine the major gene responsible for high prolificacy in the Bayanbulak sheep flock.
A small flock of Bayanbulak sheep with close genetic relationship, including 20 sheep in the sheep family with high prolificacy per lambing owned by a herdsman in Bakeshu Village, Hejing County, Xinjiang Province, is marked as Flock A (Figure 1). Up to 100 adult sheep with normal fertility were randomly selected to form flock B from 300 Bayanbulak sheep ewes in Bazhou Livestock Breeding Station in Bohu County, Xinjiang Province. All blood samples were collected from the jugular vein of the experimental sheep. About 2 ml blood sample was collected from each animal using an EDTA anticoagulant. The well-mixed blood samples were preserved at −20°C.
The main reagents used in the present study were AxyPrep-96 whole blood genomic DNA (AXYGEN, USA), Taq DNA polymerase (Qiagen Hotstar, Germany), Taq DNA ligase enzyme (NEB, UK), and dNTP (Promega, USA).
DNA extraction: Genomic DNA was extracted according to the instructions in the AxyPrep-96 whole blood genomic DNA kit, and 0.8% agarose gel electrophoresis was used to detect the integrity of the extracted genomic DNA samples.
Primers and probes: According to the sheep BMPR-IB gene sequence (AF: 312016), a pair of primers was designed to detect the FecB mutation. For the BMP15 gene sequence (NM-001114746), four pairs of primers were designed to detect the FecXI, FecXH, FecXB, FecXL, FecXG, and FecXR mutations. According to the GDF9 gene sequence (AF:078545), two pairs of primers were designed to detect the G1, G8, and FecTT mutations. Adjacent mutations in the same gene can be detected simultaneously, so the same primer sequence was designed for FecXH, FecXI, FecXL, G8, and FecTT. The above-mentioned primers were all designed by Primer Premier 5.0. The LDR probe was designed according to LDR probe design principles (Luo et al., 1996). The deletion in mutation locus (FecXR) requires a specific probe, so only FAM modification on the upstream end of primer 5’ was carried out. The primer information of PCR reaction is shown in Table 1, and the information of LDR reaction probe is shown in Table 2. All the primers and probes were synthesized by Shanghai Sangon Biological Engineering Co., Ltd.
PCR-LDR: The total volume of the PCR reaction system was 20 μl, which included 1 μl DNA (50 ng/μl) template, 2 μl 10×Buffer, 0.6 μl Mg2+ (100 mM), 2 μl dNTP (20 mM), 0.3 μl Taq DNA polymerase (5 U/μl), 4 μl 5×Q-Solution, and 0.4 μl primer mixture (5pM). The rest was ddH2O. The PCR program was as follows: pre-denaturation at 95°C for 5 min, 35 cycles of 94°C for 30 s, annealing at 53°C for 90 s, extension at 72°C for 60 s, and a final extension at 72°C for 7 min. The PCR product was tested using 3% agarose gel electrophoresis. The PCR product was diluted with isovolumetric ddH2O and used as template in the LDR detection. The total volume of the LDR reaction system was 10 μl including 1 μl 10×Buffer, 1 μl primer and probe mixture (0.5 pM/each), 0.05 μl (40U/μl) ligase (Taq DNA ligase enzyme system), 1 μl PCR product (100 ng/μl), and 6.95 μl deionized water. The LDR program was as follows: pre-denaturation at 94°C for 2 min, then 35 cycles of 30 s at 94°C, and 2 min at 50°C.
Data analysis and genotyping: In the experiment, 1 μl LDR connection product was mixed with 1 μ LABI GS 500 ROX fluorescent molecular weight standards and 1 μl deionized formamide sample. The mixture was heated at 95°C for denaturation for 2 min, placed into ice for harsh cold condition, and then underwent cataphoresis for 2.5 h in 5% polyacrylamide gel with 5 mol/L urea. GENESCAN™ 672 was used for data collection and lane line correction, as well as for size measurement of the migration section and modification of the inner molecular weight standard. Genemapper was used for data analysis and genotyping.
BMPR-IB gene: The LDR products of three different lengths were obtained in detecting BMPR-IB A746G, indicating the existence of the three genotypes.
In flock A, homozygous mutant genotype (GG) was observed 3 times giving a genotype frequency of 15%, mutation heterozygous genotype (GA) 14 times giving a genotype frequency of 70%, and wild genotype (AA) 3 times giving a genotype frequency of 15%. Mutation (G) gene frequency is 50% and the wild gene (A) frequency is 50%.
In flock B, only the wild genotype was observed, so wild gene (A) gene frequency is 100%. Mutations in the A flock was in Hardy-Weinberg equilibrium.
The independence test results of flocks A and B of Bayanbulak sheep are shown in Table 3.
Results show that the FecB polymorphism has a significant effect on the lambing performance of Bayanbulak sheep (x2 = 98.75, p<0.005).
The LDR results of the six mutation loci at the second exon of the BMP15 of Bayanbulak sheep were analyzed. The findings indicate that at FecXI, FecXH, FecXB, FecXL, FecXG, and FecXR, the lengths of LDR are 92 bp (the representative nucleotide is T), 87 bp (the representative nucleotide is C), 102 bp (the representative nucleotide is G), 99 bp (the representative nucleotide is G), 82 bp (the representative nucleotide is C), and 189 bp (lack of 17 bp deletion), respectively. At each mutation locus, the LDR products have only one length, indicating the absence of any polymorphism.
The LDR results of the G1 mutation locus of the first exon, the G8 of the second exon, and the FecTT mutation were analyzed. Results show that at the G1 mutation locus, LDR products of three lengths exist, indicating the existence of the three genotypes. At the G8 locus of GDF9, one length of 77 bp was found (the representative nucleotide is C), dedicating only homozygote genotype. At the FecTT locus of GDF9, an LDR product length of 117 bp is gained (the representative nucleotide is A), dedicating no polymorphism with only the homozygote genotype AA present.
In flock A, there are two genotypes at the G1 point mutations locus, e.g., heterozygous genotype AG was observed in one sheep giving a genotype frequency of 5%, and the wild genotype (GG) 19, with the genotype frequency of 95%. Mutation A gene frequency is 2.5%, and the wild gene G frequency is 97.5%.
In flock B, homozygous mutant genotype AA was observed 3 times giving a genotype frequency of 3%; mutation heterozygous genotype AG 8 times giving a genotype frequency of 8%; and the wild genotype GG 89 times giving a genotype frequency of 89%. Mutation A gene frequency is 7%, and the wild gene G frequency is 93%.
G1 locus, mutations in the A flock and B flock reached the Hardy-Weinberg equilibrium.
The independence test results of flocks A and B of Bayanbulak sheep are shown in Table 4.
Results show that G1 polymorphism does not affect the lambing performance of Bayanbulak sheep (x2 = 0.865, p> 0.1)
Thus far, no reports on the prolificacy gene of Bayanbulak sheep have been published. In the present paper, 10 mutation loci of 3 candidate genes of high fertility were studied for the first time. Results show that the BMPR-IB gene FecB mutation and GDF9 gene G1 mutation are present in Bayanbulak sheep. In the BMPR-IB gene FecB mutation, the GG or GA ewes produce two or more lambs per lambing. The ewes that produce two or three lambs per lambing are all GG type or GA type (Figure 1). In the control group B, no FecB mutation genotype was observed. Thus, the BMPR-IB gene FecB mutation is the major gene that influences the high fertility of Bayanbulak sheep.
According to the study of Fabre et al. (2006) the lambing number and ovulation rate are proportional to the copy number of the BMPR-IB gene. One copy of Booroola gene from the parents increases by 1.5 ova per lambing, thus producing 1 more lamb. For homozygous carriers, the increase is 3.0 ova, thus producing 1.5 more lambs (Fabre et al., 2006). Existing studies have verified the existence of the FecB mutation in some world-renowned species with high fertility such as Booroola Merino in Australia (Mulsant, 2001; Wilson, 2001), the Garole in India (Davis et al., 2002), Javanese in Indonesia (Davis et al., 2002), and Small-Tailed Han Sheep and Hu Sheep in China (Liu et al., 2003; Davis et al., 2006; Chu et al., 2007; Chu et al., 2011). The current study detected the FecB mutation in Bayanbulak sheep, making it the sixth carrier with BMPR-IB gene FecB mutation.
There are many sheep species with single mutations of reproductive capacity genes, yet only four have been reported to possess multi-locus mutations of reproductive capacity genes, including Belclare/Cambridge (Hanrahan et al., 2004) with GDF9 and BMP15 mutations, Lacaune with FecL and BMP15 locus mutations (Bodin et al., 2002), Small-Tailed Han sheep and Hu sheep with BMPR-IB and BMP15 locus mutations (Chu et al., 2007), as well as Garole with BMPR-IB and GDF9 locus mutations (Polley et al., 2009). Due to the BMPR-IB and GDF9 mutations, Bayanbulak sheep have become the sixth sheep species with two prolificacy gene mutations.
The G1 mutation of GDF9 was discovered by Hanrahan et al. (2004) in Belclare and Cambridge. This mutation leads to amino acid changes owing to the C→A mutation at 260 bp of the GDF9. Thus, the 87th amino acid residue turns into histidine from arginine. As these mutations either happen at the protease cutting site or at the undressed albumen of maturation protein, their changes do not induce the functional change of GDF9. Polley et al. (2009) reported that Garole sheep exhibit BMPR-IB and GDF9 G1 mutations. However, the relation between the mutation and the lambing number has not been revealed yet. This paper has also discovered that the GDF9 of Bayanbulak sheep has a G1 mutation with a very low mutation rate. In flock A, only one sheep was heterozygous (a lamb without lambing record); the other 19 sheep are wild type. The relevance ratio of the control group (flock B) was higher: three are mutation homozygous types and eight are mutation heterozygous types. The mutation genotype accounts for 11%. Flock B is a normal flock of Bayanbulak, so its twin lambing rate should be the mean value of the group (2% to 3%). Thus, the G1 mutation locus of GDF9 is not the major gene contributing to the large lambing number per lambing. Its relation with the twin-lamb lambing performance in Bayanbulak sheep requires further study.
The Small-Tailed Han sheep in China has high reproductive capacity with the FecB mutation, originating from the Mongolian sheep. The breed was formed after long term selection (Li-Xin and Ding-Guo, 2003). Bayanbulak sheep also evolved from Mongolian sheep. From the perspective of the breed, detecting FecB mutation from the flock with high fertility is reasonable. Thus, FecB mutation exists in Mongolian sheep with the genetic basis of high fertility, and breeding a line with high fertility is possible.
With the LDR method, two gene mutation loci with associated with fertility, namely, FecB of the BMPR-IB and G1 (FecG) of the GDF9, were found in Chinese Bayanbulak sheep. No mutation in the BMP15 gene was discovered. The mutation genotype of G1 of GDF9 has a very low ratio within the twinning flock, and the ratio is lower than that of the control group. The independence test results of flock A and B indicate that G1 of GDF9 does not affect the lambing performance of Bayanbulak sheep. Thus, G1 is not the major gene affecting the high reproductive capacity of the flock. However, the BMPR-IB FecB locus in the Bayanbulak flock with large lambing number per lambing presents corresponding gene mutations. They produce more lambs per lambing and reach a moderate polymorphic level. Independence test results of flock A and B show that the FecB polymorphic locus has a significant effect on the lambing performance of Bayanbulak sheep. Thus, this mutation is the major gene affecting the high fertility of the flock. Based on these findings, genetic resources and marker-assisted breeding technologies can accelerate the breeding of Bayanbulak sheep strains with high fertility.
ACKNOWLEDGMENTS
This study was Supported by the earmarked fund for Modern Agro-industry Technology Research System (No:CARS-39).
Figure 1
The pedigree chart of the high-prolificacy flock of Bayanbulak sheep. The gender, age, lambing performance, and FecB genotype were marked. The rectangles represent the males, whereas the circles represent the females.
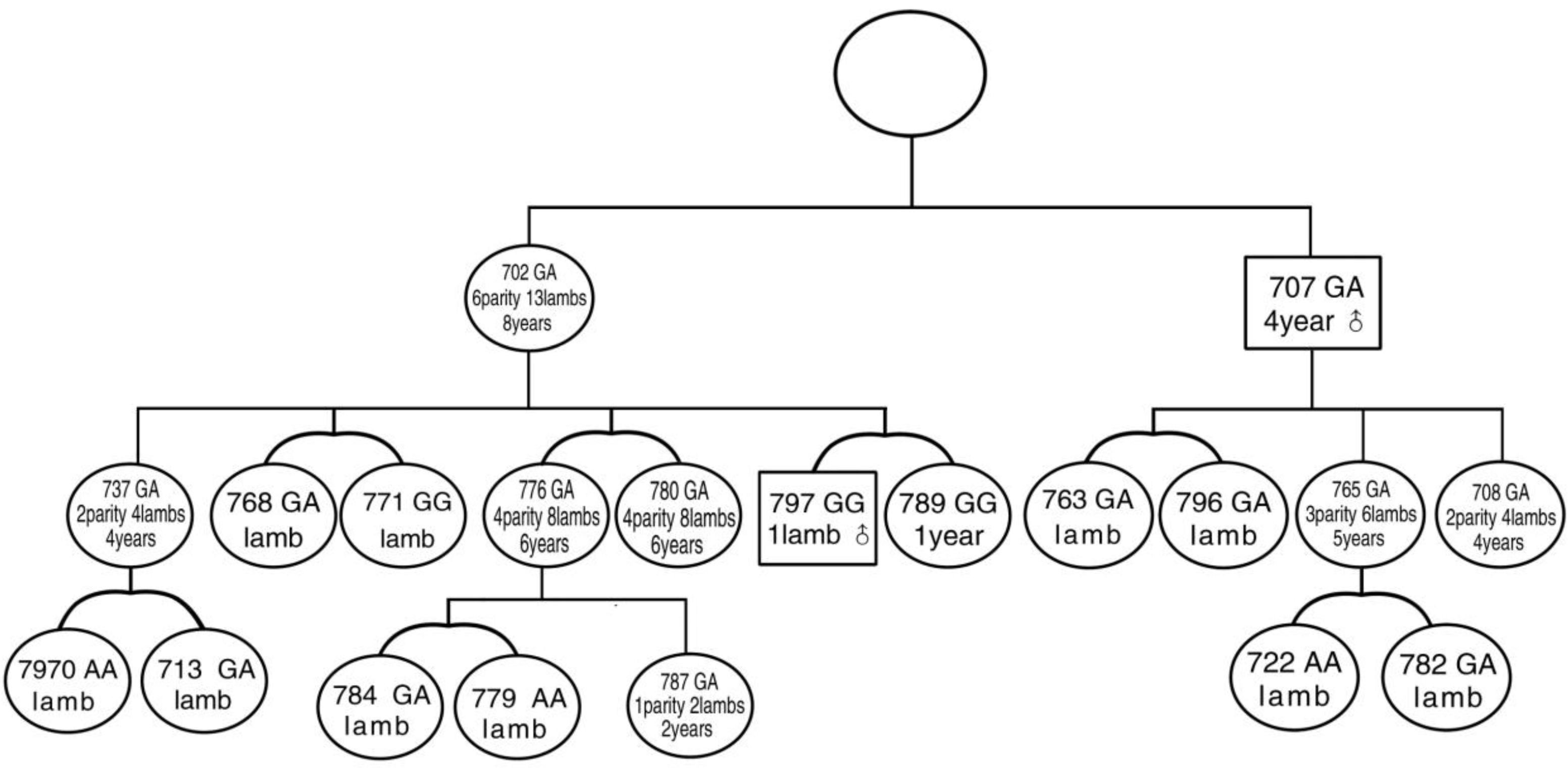
Table 1
Information of the PCR primers
Table 2
Information of the LDR probes
REFERENCES
Bodin L, Di Pasquale E, Fabre S, Bontoux M, Monget P, Ersani L, Mulsant P. 2007. A novel mutation in the bone morphogenetic protein 15 gene causing defective protein secretion is associated with both increased ovulation rate and sterility in Lacaune sheep. Endocrinology 148:393–400.


Bodin L, SanCristobal M, Lecerf F, Mulsant P, Bibe B, Lajous D, Belloc JP, Eychenne F, Amigues Y, Elsen JM. 2002. Segregation of a major gene influencing ovulation in progeny of Lacaune meat sheep. Genet Sel Evol 34:447–464.



Chu MX, Jia LH, Zhang YJ, Jin M, Chen HQ, Fang L, Di R, Cao GL, Feng T, Tang QQ, Ma YH, Li K. 2011. Polymorphisms of coding region of BMPR-IB gene and their relationship with litter size in sheep. Mol Biol Rep 38:4071–4076.


Chu MX, Liu ZH, Jiao CL, He YQ, Fang L, Ye SC, Chen GH, Wang JY. 2007. Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep (Ovis aries). J Anim Sci 85:598–603.


Davis GH, Balkrishnan L, Ross IK, Wilson T, Galloway SM, Lumsden BM, Hanrahan JP, Mullen M, Mao XZ, Wang GL, Zhao ZS, Zeng YQ, Robinson JJ, Mavrogenis AP, Papachristoforou C, Peter C, Baumung R, Cardyn , Baujenane I, Cockett NE, Eythorsdottir E, Arranz JJ, Notter DR. 2006. Investigation of the Booroola (Fec B) and Inverdale (Fec X I) muatation in 21 prolific breeds and strains of sheep samples in 13 countries. Anim Reprod Sci 92:87–96.


Davis GH, Galloway SM, Ross IK, Gregan SM, Ward J, Nimbkar BV, Ghalsasi PM, Nimbkar C, Gray GD, Subandriyo , Inounu I, Tiesnamurti B, Martyniuk E, Eythorsdottir E, Mulsant P, Lecerf F, Hanrahan PJ, Bradford GE, Wilson T. 2002. DNA Tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biol Reprod 66:1869–1874.


Fabre S, Pierre A, Mulsant P, Bodin L, DiPasquale E, Persani L, Monget P, Monniaux D. 2006. Regulation of ovulation rate in mammals: contribution of sheep genetic models. Reprod Biol Endocrinol 4:20



Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Jennifer JL, Jokiranta S, McLaren RJ, Luiro K, Dodds KG, Montgomery GW, Beattie AE, Davis GH, Ritvos O. 2000. Mutations in an oocyte-derived growth factor gene (BMP15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat Genet 25:279–283.


Hanrahan PJ, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, Galloway SM. 2004. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries). Biol Reprod 70:900–909.


Juengel JL, Hudson NL, Heath DA. 2002. Growth differentiation factor9 and bone morphogenetic protein 15 are essential for ovarian follicular development in sheep. Biol Reprod 67:1777–1789.


Du LX, Cao DG. 2003. Studies on microsatellite and RAPD markers in little-tailed han sheep. Acta Genet Sin 30:1041–1044.
Linda N, Stephen CB, Ricardo PW, Christian B, Lars-Erik H, Stewart MR, Alan SM. 2009. Homozygosity for a single base-pair mutation in the oocyte specific GDF9 gene results in sterility in Thoka sheep. Reproduction 138:921–933.


Luo J, Bergstorm DE, Barany F. 1996. Improving the fidelity of thermos thermophilus DNA ligase. Nucleic Acids Res 24:3071–3078.



Liu SF, Jiang YL, Du LX. 2003. Studies of BMPRIB and BMP15 as candidate genes for fecundity in little tailed han sheep. Yi Chuan Xue Bao (Acta Genetica Sinica) 30:755–760.

Martinez-Royo A, Jurado JJ, Smulders JP, Marti JI, Alabart JL, Roche A, Fantova E, Bodin L, Mulsant P, Serrano M. 2008. A deletion in the bone morphogenetic protein 15 gene causes sterility and increased prolificacy in Rasa Aragonesa sheep. Anim Genet 39:294–297.


Monteagudo LV, Ponz R, Tejedor MT, Lavina A, Sierra I. 2009. A 17 bp deletion in the bone morphogenetic protein 15 (BMP15) gene is associated to increased prolificacy in the Rasa Aragonesa sheep breed. Anim Reprod Sci 110:139–146.


Moore RK, Shimasaki S. 2005. Molecular biology and physiological role of the oocyte factor, BMP-15. Mol Cell Endocrinol 234:67–73.


Otsuka F, Yamamoto S, Erickson GF. 2001. Bone morphogenetic protein-15 inhibits follicle-stimulating hormone (FSH) action by suppressing FSH receptor expression. Biol Chem 276:11387–11392.

Otsuka FZ, Yao T, Lee S. 2000. Bone morphogenetic protein-15 Identification of target cells and biological functions. Biol Chem 275:39523–39528.

Mulsant P, Lecerf F, Fabre S, Schibler L, Monget P, Lanneluc I, Pisselet C, Riquet J, Monniaux D, Callebaut I. 2001. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc Natl Acad Sci 98:5104–5109.



Polley S, De S, Batabyal S, Kaushik R, Yadav P, Arora JS, Chattopadhyay S, Pan S, Brahma B, Datta TK, Goswami SL. 2009. Polymorphism of fecundity genes (BMPR1B, BMP15 and GDF9) in the Indian prolific Black Bengal goat. Small Rumin Res 85:122–129.

Polley S, De S, Brahma B. 2009. Polymorphism of BMPR1B, BMP15 and GDF9 fecundity genes in prolific Garole sheep. Trop. Anim. Health Prod. 10.1007/s11250-009-9518-1

Wilson T, Wu XY, Juengel JL, Ross IK, Lumsden JM, Lord EA, Dodds KG, Walling GA, McEwan JC, O’Connell AR. 2001. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol Reprod 64:1225–1235.


- TOOLS





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





