Characterization of Insertional Variation of Porcine Endogenous Retroviruses in Six Different Pig Breeds
Article information
Abstract
Pigs may need to be exploited as xenotransplantation donors due to the shortage of human organs, tissues and cells. Porcine endogenous retroviruses (PERVs) are a significant obstacle to xenotransplantation because they can infect human cells in vitro and have the potential for transmission of unexpected pathogens to humans. In this research, 101 pigs, including four commercial breeds (23 Berkshire, 13 Duroc, 22 Landrace and 14 Yorkshire pigs), one native breed (19 Korean native pigs) and one miniature breed (10 NIH miniature pigs) were used to investigate insertional variations for 11 PERV loci (three PERV-A, six PERV-B and two PERV-C). Over 60% of the pigs harbored one PERV-A (907F8) integration and five PERV-B (B3-3G, B3-7G, 742H1, 1155D9 and 465D1) integrations. However, two PERV-A loci (A1-6C and 1347C1) and one PERV-B locus (B3-7F) were absent in Duroc pigs. Moreover, two PERV-C loci (C2-6C and C4-2G) only existed in Korean native pigs and NIH miniature pigs. The results suggest that PERV insertional variations differ among pig breeds as well as among individuals within a breed. Also, the results presented here can be used for the selection of animals that do not have specific PERV integration for xenotransplantation research.
INTRODUCTION
Livestock species have been widely used for the biomedical researches, especially for chickens and pigs (Matsumoto et al., 2011; Robb et al., 2011; Luo et al., 2012). Xenotransplantation using porcine cells, tissues and organs has focused on biomedical aspects, with the aim of decreasing wait times for patients who require an allotransplantation. Pigs are considered as the best alternative xenotransplant organ donor for humans because of the anatomical and physiological similarities of pig organ size with humans, economic benefit of their quick reproductive capability and the relatively decreased ethical concerns with pig sacrifice compared with primates (Lee and Moran, 2001). Also, Weiss et al. (1998) reported that genetically engineered pigs can provide donor material that successfully evades xenograft rejection by the recipient immune system. Practically, however, xenotransplantation still is fraught with immunological problems and carries the potential risk of transmission of zoonotic microbial pathogens from the pig donor to the human recipient.
Porcine endogenous retroviruses (PERVs) are one of the important obstacles in xenotransplantation because they may carry the potential risk of cross-species transmission to humans. Based on their envelope protein types, PERVs are classified as PERV-A, PERV-B and PERV-C. Once acquired, inheritance can occur in a Mendelian fashion (Patience et al., 1997; Blusch et al., 2002). Also, more than 50 copies of PERVs are integrated within the genome of all pig breeds (Le Tissier et al., 1997; Akiyoshi et al., 1998; Jung et al., 2010). Especially, PERV-A and PERV-B are human-tropic viruses that can infect a wide range of human and other mammalian cells in vitro, indicating that they are a potential risk factor for xenotransplantation (Patience et al., 1997; Takeuchi et al., 1998; Wilson et al., 2000; Specke et al., 2001, 2002), whereas ecotropic PERV-C viruses only infect pig cells in vitro (Wilson et al., 2000; 2008). Recently, Denner (2008) discussed the risks of PERV-A and PERV-C recombinant viruses, which were capable of infecting human cells in xenotransplantation. Therefore, many researchers have been trying to prevent transmission of PERVs from pig cells to human cells using knock-out of active PERVs in the pig genome and knock-down of PERV expression by PERV-specific small interfering RNA (siRNA) (Dieckhoff et al., 2008; Butler et al., 2009). It has proven challenging to generate PERV-C free pig breeds to reduce the risk of recombinant PERV-A and PERV-C transmission to humans. Also, there were many variations in PERV integration sites among pig breeds such as Westran pig, Large White pig, Korean native pig and NIH miniature pig (Lee et al., 2002; Jung et al., 2010; Yu et al., 2012).
The aim of this study was to investigate PERV insertional variations in different pig breeds using 11 selected PERV loci from Korean native pig (KNP) and National Institutes of Health (NIH) miniature pig bacterial artificial chromosome (BAC) libraries. The results could provide valuable information for the selection of animals that do not have specific PERV integration for xenotransplantation studies.
MATERIALS AND METHODS
Experimental animals and development of PERVs
Previously, PERV positive BAC clones were screened from KNP and NIH miniature pig BAC libraries in Korea (Jung et al., 2010; Yu et al., 2012). Of these, 11 non-redundant PERV clones were used for sequencing of PERV flanking DNA. To investigate the PERV insertional variations, 101 pigs from six different pure-bred pig breeds (22 Landrace pigs, 13 Duroc pigs, 23 Berkshire pigs, 14 Yorkshire pigs, 10 NIH miniature pigs and 19 Korean native pigs) were obtained from Gyeongsang National University (GNU), Korea. Genomic DNAs from these pigs were extracted from blood using standard protocols. The genomic DNA concentrations were measured using a NanoDrop 2000C spectrophotometer (Thermo Scientific, USA).
Investigation of PERV insertional variations in six different pig breeds
To investigate insertional variations in each PERV locus, PERV insertion-specific primers were designed from the 5’ and 3 ’ genomic flanking sequences obtained from six PERV positive BAC clones (A1-6C, B3-3G B3-7F, B3-7G C2-6C and C4-2G) in NIH miniature pig (Yu et al., 2012). Also, five previously well-characterized PERV BAC clone sequences (465D1, 742H1, 907F8, 1155D9 and 1347C1) from KNP (Jung et al., 2010) were used as PERV insertion identification markers. Internal PERV control primers were designed to amplify conserved regions between 5’ long terminal repeat (LTR) and 3’ env gene (Table 1). To confirm the PERV integrations, both the 5’ and 3’ flanking regions for each PERV were amplified by polymerase chain reaction (PCR). PCR amplification was carried out in a 25 μl total volume containing 20 ng of template gDNA, 10×PCR reaction buffer (Genetbio, Korea), 2.5 mM of each dNTP (Genetbio, Korea), 10 pmol of each primer and 1 unit of Taq polymerase (Genetbio, Korea). The cycling conditions included an initial incubation of 5 min at 94°C followed by 35 cycles of 94°C for 30 s, 60 to 65°C for 30 s, 72°C for 45 s, and the final elongation was performed at 72°C for 5 min using a PTC-200 Programmable Thermal Controller (MJ Research, USA). The PCR products were electrophoresed in a 2% standard 1X TAE agarose gel stained with ethidium bromide (EtBr). Direct sequencing reactions of the PCR products were performed using the ABI PRISM BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) with the sequencing primers indicated in Table 1. The products were analyzed on an ABI 3730XL automated DNA analyzer (Applied Biosystems, USA).
RESULTS AND DISCUSSION
There are variations in PERV copy numbers among different pig breeds (Allan et al., 1999; Lee et al., 2002; Jung et al., 2010; Yu et al., 2012). Moreover, Moalic et al. (2009) suggested that most of the PERV integration sites in infected human cells (HEK-293) are located within a 5 kb region of CpG islands, transcriptional regulation and transcription sites (Patience et al., 1997; Park et al., 2010). To investigate the PERV insertional variations among different pig breeds, previously reported six PERV positive BAC clones (one PERV-A positive BAC clone: A1-6C; three PERV-B positive BAC clones: B3-3G B3-7F and B3-7G; two PERV-C positive BAC clones: C2-6C and C4-2G), identified in the NIH miniature pig BAC library, were used for the PERV variation studies (Yu et al., 2012). PERVs in these clones were subjected to full-length PERV nucleotide sequencing of both the 5’ and 3’ genomic flanking sequences. These sequences were used for designing primers for screening PERV site-specific insertional variations (Figure 1, Table 1). In addition, previously detected 5’ and 3’ PERV genomic flanking sequences of five PERV positive BAC clones (two PERV-A positive BAC clones: 907F8 and 1347C1; three PERV-B positive BAC clones: 465D1, 742H1 and 1155D9), originating from the KNP BAC library, were used to investigate insertional variations in each PERV locus (Jung et al., 2010). Therefore, in this study, 11 PERV locations (three PERV-A, six PERV-B and two PERV-C) were investigated for the variations of 101 pigs from six different pig breeds, including 22 Landrace pigs, 13 Duroc pigs, 23 Berkshire pigs, 14 Yorkshire pigs, 10 NIH miniature pigs and 19 Korean native pigs (Figure 2, Table 2). In order to investigate the replication competent PERVs, open reading frame (ORF) structure has been observed for the selected 11 PERV positive BAC clones (Figure 3). The data indicated that 3 out of 11 clones (1347C1, B3-7F and C2-6C) have replication competent PERVs which can make full PERV particles (Yu et al., 2011).
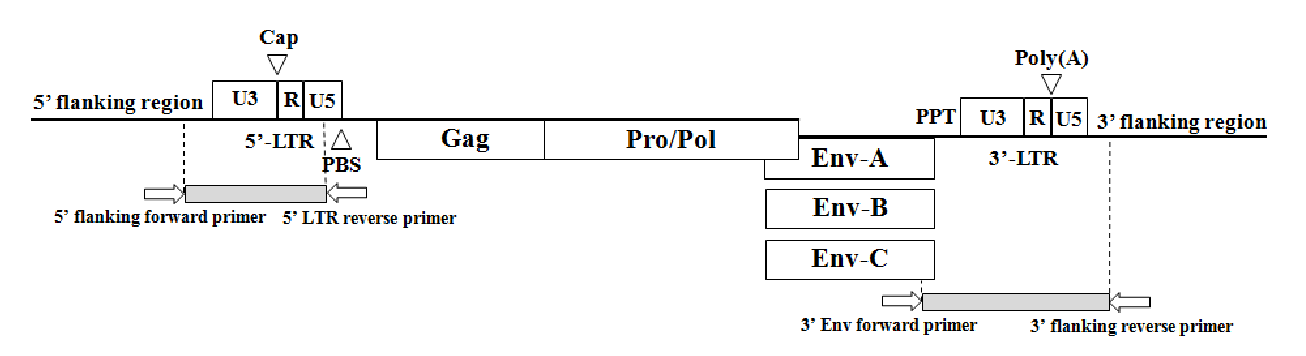
Structure of the porcine endogenous retrovirus (PERV) genome and a schematic diagram indicating the primer locations for identifying PERV site specific insertional variations. Also, open boxes indicated genes and open reading frames. Abbreviations: Cap = Transcriptional start site; PBS = Primer binding site; PPT = Polypurine tract; Poly(A) = Poly(A) addition site.
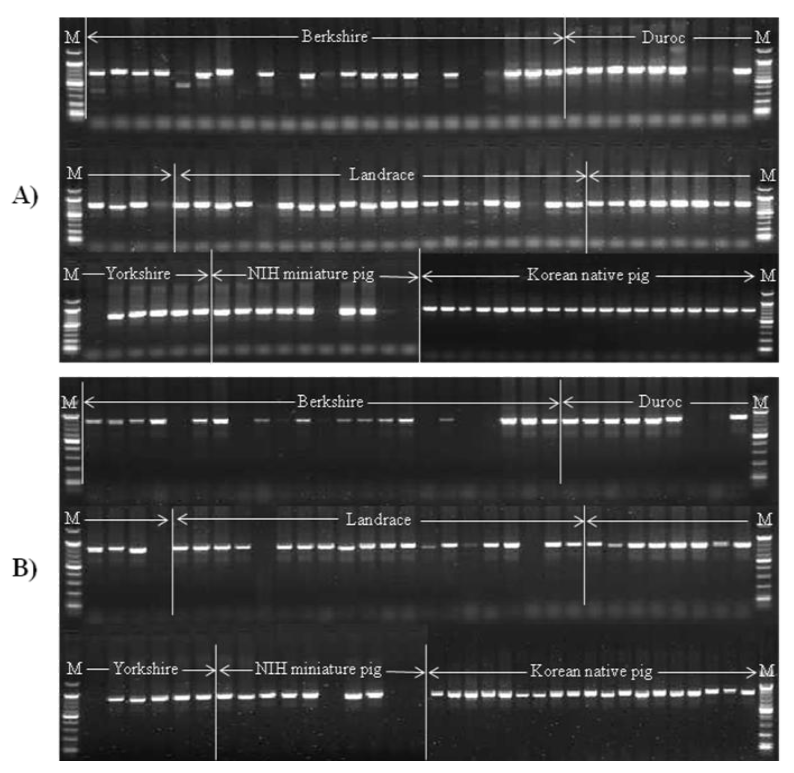
Representative examples of identified insertional variations at 907F8 locus in six different pig breeds. A) The primers were designed from 5’ flanking region to 5’ LTR region. B) The primers were designed from 3’ envelop region to 3’ flanking region. The breeds consisted of 23 Berkshire pigs, 13 Duroc pigs, 22 Landrace pigs, 14 Yorkshire pigs, 10 NIH miniature pigs and 19 Korean native pigs. The amplified PCR product size is indicated in Table 1 (M: 100 bp DNA ladder marker).
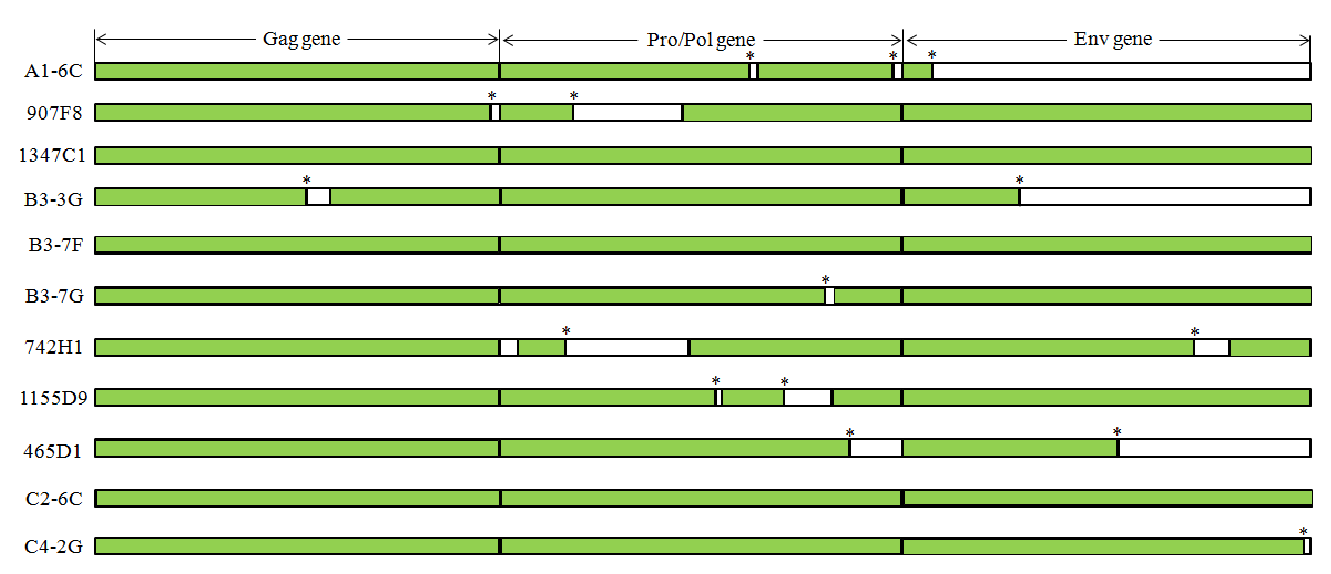
Open reading frame (ORF) structure of 11 PERV positive clones selected from Korean native pig and NIH miniature pig BAC libraries. Three clones (1347C1, B3-7F and C2-6C) have intact ORFs, indicating a high possibility of making perfect virus particles (Yu et al., 2011). Star mark (*) indicates a stop codon.
All the NIH miniature pigs used in this research had an A1-6C PERV-A locus, which was located on SSC13. Assessment of this locus in KNPs revealed an insertional rate of 84.2%. On the other hand, the Al-6C locus in Landrace and Berkshire pigs showed a very low insertion rate of 13.6% and 17.4%, respectively. Also, this PERV integration was not observed in all Duroc and Yorkshire pigs. A 907F8 PERV-A locus, located on SSC17, has been found in all KNPs. In case of Yorkshire and Landrace pigs, this locus displayed a very high insertion rate of 92.9% and 90.9%, respectively. Also, more than 70.0% of Berkshire, Duroc and NIH miniature pigs had this locus. Jung et al. (2010) reported that chromosomal location of a 1347C1 PERV-A locus could not be determined using the SCH and IMpRH panels because the identified BAC-end sequences of 1347C1 PERV clone contained a large portion of repetitive sequences. This 1347C1 locus has not been found in all Duroc pigs investigated. However, the other five pig breeds had insertion rates of 30.0% to 58.0%, indicating insertional variations among breeds. The three identified PERV-A loci were variable in most of the six pig breeds. Especially, PERVs were unfixed in the pig genome of Duroc at both the A1-6C and 1347C1 loci.
In this study, six loci (B3-3G, B3-7F, B3-7G, 465D1, 742H1 and 1155D9) were used for identifying PERV-B specific insertional variations. Three PERV-B loci (B3-3G, B3-7F and B3-7G) were selected from the NIH miniature pig BAC library. All NIH miniature pigs had the B3-3G locus, located on SSC4. Duroc pigs had the locus that corresponded to a high PERV insertional rate of 76.9%, followed by Landrace pigs with a rate of 59.1%, KNPs with a rate of 57.9%, Yorkshire pigs with a rate of 50.0% and Berkshire pigs with a rate of 17.4%. In contrast to the B3-3G locus, the B3-7F PERV integration was very low in Duroc (0.0%) and NIH miniature pigs (20.0%). Also, fewer than 32.0% of Berkshire, Landrace and NIH miniature pigs possessed this locus. However, KNPs and Yorkshire pigs had a relatively high insertional rate of 73.7% and 92.9% for the B3-7F locus, respectively. The B3-7G locus, located on SSC3, was found in all Duroc pigs, representing the highest PERV insertional rate. Berkshire pigs had the locus with a high insertional rate of 91.3%, followed by NIH miniature pigs with a rate of 90.0%, KNPs with a rate of 78.9%, Landrace pigs with a rate of 72.7% and Yorkshire with a rate of 71.4%. Also, all PERV-B loci except the 1155D9 locus, which was fixed in all animals used in this study, variably existed in the pig genome.
Three other PERV-B loci (465D1, 742H1 and 1155D9), which originated from the KNP BAC library, displayed varied insertional rates among the breeds. The 742H1 locus, located on SSC2, was identified in all NIH miniature pigs. The integrated rates of this 742H1 PERV locus in Duroc (84.6%), Berkshire (82.6%) and KNPs (57.9%) were relatively higher than those in Landrace (36.4%) and Yorkshire pigs (21.4%). Also, the 1155D9 locus, located on SSC7, was identified in all pig breeds with PERV insertional rates of 90.9%, 89.5%, 60.9%, 57.1%, 20.0% and 7.7% in Landrace pigs, KNPs, Berkshire pigs, Yorkshire pigs, NIH miniature pigs and Duroc pigs, respectively. In case of the 465D1 locus, all Duroc and KNP individuals had the locus. Also, Landrace, Yorkshire and Berkshire breeds had the locus that corresponded to comparatively high insertional rates of 91.0%, 78.6% and 78.3%, respectively. However, the 465D1 locus was not detected in all NIH miniature pigs.
Recently, chromosomal locations of four PERV-C positive BAC clones (C1-10G, C2-6C, C3-6F and C4-2G) were identified in NIH miniature pigs (Yu et al., 2012). In this study, two clones (ID: C2-6C and C4-2G) of the previously-identified four PERV-C positive BAC clones were selected for characterizing PERV-C specific insertional variations. Especially, Fujimura et al. (2008) reported that PERV-C types were only found in a few pig breeds such as Landrace (16%; 8/50), Large White (21%; 9/43), Duroc (52%; 26/50), Berkshire (68%; 129/191), miniature pigs (83%; 5/6) and genetically modified triple cross-breed (LWD) pigs (100%; 36/36). However, two PERV-C loci, the C2-6C locus located on SSC13 and the C4-2G locus located on SSC13, were not found in all Berkshire, Duroc, Landrace and Yorkshire pigs used in this study. On the other hand, the two selected PERV-C loci from NIH miniature pig BAC library indicated that each had the same integrated rates of 73.7% in KNPs. In NIH miniature pigs, the integration rate (40%) of PERVs at the C2-6C locus was relatively lower than that (90%) of the C4-2G locus.
In order to confirm the absence of PERVs in the PCR negative animals at each PERV locus, we have tried to amplify full-length PERVs with primers designed from 5’ and 3’ flanking regions to amplify the flanking sequence from PERV negative animals. These primers were also used to determine whether the PCR positive animals are homozygous or heterozygous for each PERV locus. The data indicated that nine PERV loci (A1-6C, 907F8, B3-7F, B3-7G, 742H1, 1155D9, 465D1, C2-6C and C4-2G) were well amplified for genotyping of homozygous and heterozygous animals. On the other hand, 18 animals (17.8%) of 1347C1 and 14 animals (13.9%) of B3-3G PERV loci were not genotyped well which may indicate the PCR was not amplified due to the mutations in the primer binding sites (Table 2).
In conclusion, we have investigated 11 PERV insertional variations in six different pig breeds. Even though we studied small number of pigs in each breed, more than 60% of the investigated animals had one PERV-A (907F8) integration and five PERV-B (B3-3G, B3-7G, 742H1, 1155D9 and 465D1) integrations. However, two PERV-A loci (A1-6C and 1347C1) and one PERV-B locus (B3-7F) were absent in Duroc pigs. Especially, two PERV-C loci (C2-6C and C4-2G) only existed in Korean native pigs and NIH miniature pigs. This indicates that they have a relatively high probability of producing PERV-A and PERV-C recombinant PERVs in xenotransplantation. Based on these results, we conclude that PERV integrations differ among breeds as well as among individuals. The results of the present study can be used for the efficient selection of animals that do not have specific PERV integration, especially for xenotransplantation studies.
ACKNOWLEDGEMENTS
This work was supported by a grant (No. 20070401034031) from BioGreen 21 program, Rural Development Administration, Republic of Korea and by a grant (No. 20100023352) from the National Research Foundation of Korea, Republic of Korea. Also, authors give special thanks to Animal Genomics and Bioinformatics Division in National Institute of Animal Science, RDA, for accessing BAC libraries.

