Molecular characterization and expression pattern of a novel Keratin-associated protein 11.1 gene in the Liaoning cashmere goat (Capra hircus)
Article information
Abstract
Objective
An experiment was conducted to determine the relationship between the KAP11.1 and the regulation wool fineness.
Methods
In previous work, we constructed a skin cDNA library and isolated a full-length cDNA clone termed KAP11.1. On this basis, we conducted a series of bioinformatics analysis. Tissue distribution of KAP11.1 mRNA was performed using semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis. The expression of KAP11.1 mRNA in primary and secondary hair follicles was performed using real-time PCR (real-time polymerase chain reaction) analysis. The expression location of KAP11.1 mRNA in primary and secondary hair follicles was performed using in situ hybridization.
Results
Bioinformatics analysis showed that KAP11.1 gene encodes a putative 158 amino acid protein that exhibited a high content of cysteine, serine, threonine, and valine and has a pubertal mammary gland) structural domain. Secondary structure prediction revealed a high proportion of random coils (76.73%). Semi-quantitative RT-PCR showed that KAP11.1 gene was expressed in heart, skin, and liver, but not expressed in spleen, lung and kidney. Real time PCR results showed that the expression of KAP11.1 has a higher expression in catagen than in anagen in the primary hair follicles. However, in the secondary hair follicles, KAP11.1 has a significantly higher expression in anagen than in catagen. Moreover, KAP11.1 gene has a strong expression in inner root sheath, hair matrix, and a lower expression in hair bulb.
Conclusion
We conclude that KAP11.1 gene may play an important role in regulating the fiber diameter.
INTRODUCTION
Cashmere is a heterogeneous fleece mainly composed of unmedulated cashmere and medulla developed wool, which are, respectively, generated by the secondary and the primary hair follicles [1]. After the wool follicles become mature and active, it undergoes three regular periods termed anagen, catagen and telogen in a yearly cycle [2]. Cashmere characteristics are distinctly different depending on the region and goat species [3].
Cashmere is the derivative of the secondary hair follicles [1] and is the inner hair of the goat. Keratins and keratin-associated proteins (KAPs) are the main structural proteins of the hair, which determine the essential characteristic of cashmere and wool [4]. KAP and keratin intermediate filament (IF) are the major components of hair [5]. KAPs are structural proteins used for encoding hair fiber. More than 100 KAP genes that have been grouped into 27 KAP families have been identified in mammalian species [6]. In sheep all the KAP genes that have been identified have between two and nine alleles [4]. KAPs can be subdivided into three groups based on their chemical properties: i) The high sulfur KAPs (<30 mol % cysteine content) consisting of the KAP1–KAP3, KAP10–KAP16, and KAP23–KAP27 families; ii) the ultrahigh sulfur KAPs (>30 mol % cysteine content) comprising the KAP4, KAP5, KAP9, and KAP17 families; and iii) the high glycine tyrosine KAPs consisting of the KAP6–KAP8 and KAP18–KAP22 families [6,7]. Separate multi-gene families have been shown to code for each of these groups [8]. The IF proteins form the skeleton of hair, which has a content and structure that are relatively stable. But the content and structure of KAPs differs greatly in different species’ hair. So KAPs may play a major role in the quality of hair. To further study the influence of KAPs on hairs, and to lay a foundation for breeding superior quality cashmere goats. We cloned the full-length cDNA sequence of KAP 11.1, analyzed the cDNA and amino acid sequences, and established an evolutionary tree. Then we analysed the structural domains and hydrophobicity, forecast secondary structure, including transmembrane domains, phosphorylation sites, signal peptides, and the subcellular localization. Reverse transcription-polymerase chain reaction (Semi-quantitative RT-PCR) was used to detect the expression of the KAP11.1 gene in all internal organs. Real-time RT-PCR was used to detect the levels of KAP11.1 gene expression in the primary and secondary hair follicles during anagen, catagen, and telogen. All these findings lay a solid foundation for further studying the mechanism regulating the KAP11.1 gene.
MATERIALS AND METHODS
Tissue samples collection and hair follicle’s separation
The research protocols in this study were approved by the Animal Experiment and Care Committee of Liaoning Normal University. Liaoning cashmere goats were randomly selected from cashmere goat farm at Wafangdian, Dalian, China. Tissues including heart, liver, spleen, lungs andkidney were collected from goats during anagen after slaughter. Tissue samples were immediately frozen in liquid nitrogen and stored at –80°C. We separated the primary and secondary hair follicles during anagen and catagen, and extracted total RNA. First, the skin at anagen and catagen was cut into a 0.5 cm wide strip, exposing the complete hair follicle bulb, and then the subcutaneous fat was removed with a dissecting needle. Forceps were used to slowly separate the hair follicle tissues along the direction of growth, and then the needle was used to remove surrounding tissues. Undamaged hair follicles were placed into the culture medium [9]. The Trizol was used to extract total RNA.
Sequence and bioinformatics analysis of the Liaoning cashmere goat KAP11.1
In previous work, we constructed a skin cDNA library and isolated a full-length cDNA clone termed KAP11.1. The sequence was analyzed by the BLASTN/BLASTX to identify DNA/protein homologues and possible functions. The open reading frame was identified using NCBI’s ORF finder. The evolutionary tree was constructed using Mega3.2. The amino acid composition, molecular weight, and isoelectric point were analyzed using ProParam (http://web.expasy.org/protparam/). Protein domain prediction was performed using Pfam (http://pfam.xfam.org/). The secondary structure of KAP11.1 was predicted using PredictProtein (http://www.predictprotein.org/). The hydrophobicity was analyzed by ProtScale (http://web.expasy.org/protscale/). The transmembrane domain prediction was performed using the TMHMM program (http://www.cbs.dtu.dk/services/tmhmm/). The phosphorylation site prediction was performed by NetPhos program (http://www.cbs.dtu.dk/services/NetPhos/). The deduced signal peptide was identified using SignalP (http://www.cbs.dtu.dk/services/SignalP/). The subcellular localization prediction was carried out using EpiLoc program (http://epiloc.cs.queensu.ca/).
Tissue distribution of the Liaoning cashmere goat KAP11.1 mRNA
Tissue distribution of KAP11.1 mRNA was performed using semi-quantitative RT-PCR analysis.
Total RNA was extracted from skin, heart, liver, spleen, lungs, and kidney during anagen. Reverse transcription was performed following the RT-PCR (Takara, DaLian, China) Kit instructions. cDNA products were preserved in –20°C. Specific primers were designed to amplify KAP11.1 and β-actin cDNA fragments from goat, and the RT products were used as template. The reaction system (25 μL) was as follows: 29 Master Mix, 12.5 μL; RT products, 2 μL; the upstream and downstream primers, 1 μL; deionized water, 8.5 μL. The protocol was an initial denaturation at 94°C for 3 min, then 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 60 s, followed by 72°C for 5min. Agarose gel electrophoresis and a UV spectrophotometer were used to detect the PCR products. KAP11.1 mRNA was detected in different tissues while β-actin as a loading control. The primers used are listed in Table 1.
Expression of the Liaoning cashmere goat KAP11.1 mRNA in primary hair follicles and secondary hair follicles
The expression of KAP11.1 mRNA in primary and secondary hair follicles was performed using real-time polymerase chain reaction (real-time PCR) analysis. Total RNA was obtained from primary and secondary hair follicles and from anagen and catagen skin using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Agarose (1%) gel electrophoresis and a UV spectrophotometer were used to detect the RNA purity and concentration. RNA was stored as an aqueous solution at –80°C. Prior to real-time PCR, the RNA samples were treated with DNAseI. The purified total RNA was again analyzed for purity and concentration. Reverse transcription was performed following the RT-PCR (Takara, DaLian) Kit instructions. cDNA products were preserved in –20°C. The primers used are listed in Table 1.
Reaction system was a total of 20 μL, including 10 μL of SYBR Premix Ex TaqII (29), 0.4 μL of upstream and downstream primers, 1 μL of RT product, and 8.2 μL of dH2O. The PCR conditions applied were a 95°C pre-degeneration for 10 s followed by 40 cycles of 95°C denaturation for 5 s, 60°C annealing for 30 s, and 72°C extension for 60s. The β-actin gene served as a reference, and each sample run was repeated three times.
Expression location of the Liaoning cashmere goat KAP11.1 mRNA in primary hair follicles and secondary hair follicles
The Expression location of KAP11.1 mRNA in primary and secondary hair follicles was performed using in situ hybridization. After cloning and sequencing, RT-PCR products of KAP11.1 gene were repeated purified, generated the KAP11.1 cDNA product concentration for 104.5 ng/μL, which was used as a Digoxigenin Labeled template. Label was performed according to digoxigenin kit instruction (Roche, Germany), and the prepared probes were stored in –20°C refrigerator. Tissue sections of in situ hybridization were via dewaxing, rehydration, proteinase K digestion, dry, and then pre-hybridization 1 h with 50% deionized formamide, 42°C hybridization overnight, the hybridized action was placed in turn in 29 standard saline citrate (SSC) wet box, 19 SSC, 0.259 SSC washing, after hybridization adding antibody dilution according to the 1:500, combined with antibodies order to immunological detection, using Tetranitroblue tetrazolium chloride/5-Bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) color, then neutral resin Seal Sheet. Observe by microscope and take photos.
Statistical analysis
Statistical analyses were accomplished with SPSS 17.0 (SPSS Inc., Chicago, IL, USA). Data was expressed as mean±standard deviation of three independent experiments. Less than 0.05 p value was considered thought statistically significant for all tests.
RESULTS
Sequence and bioinforematics analysis of the Liaoning cashmere goat KAP11.1
KAP11.1 gene cDNA is 940 bp and contains an intact open reading frame of 480 bp encoding a putative 159 amino acid protein. There was only one methionine codon. Additionally, the cDNA clone contained a conventional polyadenylation site 133 bp downstream of the coding region (Figure 1).

The Liaoning cashmere goat’s KAP11.1 gene cDNA sequence and amino-acid translation analysis. The corresponding amino acid sequence is below the nucleotide sequence. The circled amino acid signifies the putative in-frame initiation of translation codon. The waved line at the end of the cDNA sequence denotes the polyadenylation signal. The dotted line below the cDNA sequence shows PMG regions. KAP, Keratin-associated protein; PMG, pubertal mammary gland.
The deduced amino acid sequence of Liaoning cashmere goat’s KAP11.1 gene showed high homology with KAP11.1 gene of bos Taurus (NM_001080740.1), homo sapiens (NM_175858.2), mus musculus (NM_001113406.1), and rattus norvegicus (XM_ 002727860.1) at 91%, 77%, 66%, and 75%, respectively (Figure 2). The putative proteins were shown to be rich in cysteine, glycine, serine, threonine, tyrosine and valine; however, they have a relatively low amount of histidine, lysine and methionine. In addition, the amount of lysine is generally low and the amount of glycine and serine is generally high in all putative proteins as compared with other amino acids. What’s more, KAP11.1 contains relatively high amount of threonine, valine and relatively low amount of metnionine in comparison with other putative proteins (Table 2). To study the evolutionary relationship of Liaoning cashmere goat’s KAP11.1 gene with other mammalian KAP11.1 genes, we constructed a phylogenetic tree by aligning the amino acid sequences. KAP11.1, together with KAP3.1 and KAP3.2, formed a cluster from the same branch and had high homology (Figure 3A). Additionally, KAP11.1 of Liaoning cashmere goat had the highest homology with cattle and the lowest homology with rat, which is in accordance with the analysis of homologous amino acid sequence (Figure 3B).

Comparison of KAP11.1 amino acid sequences in five species. An alignment of KAP11.1 amino acid sequences is presented in the single letter code with gaps introduced to maximize the amino acid alignment (dashes). Asterisks below the alignment indicate sequence identity. Colons denote conservative substitutions and dots show non-conservative substitutions. Bold type highlights amino acids with strikingly similar/dissimilar mol% values. KAP, Keratin-associated protein.
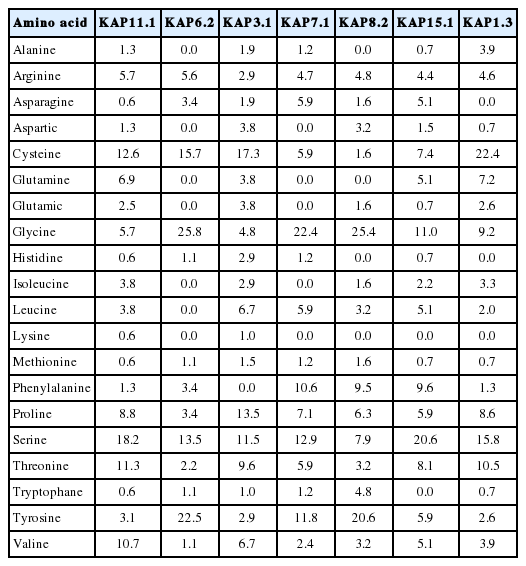
Comparison of KAP11.1 amino-acid composition (mol%) with other keratin-associated proteins of goat or sheep
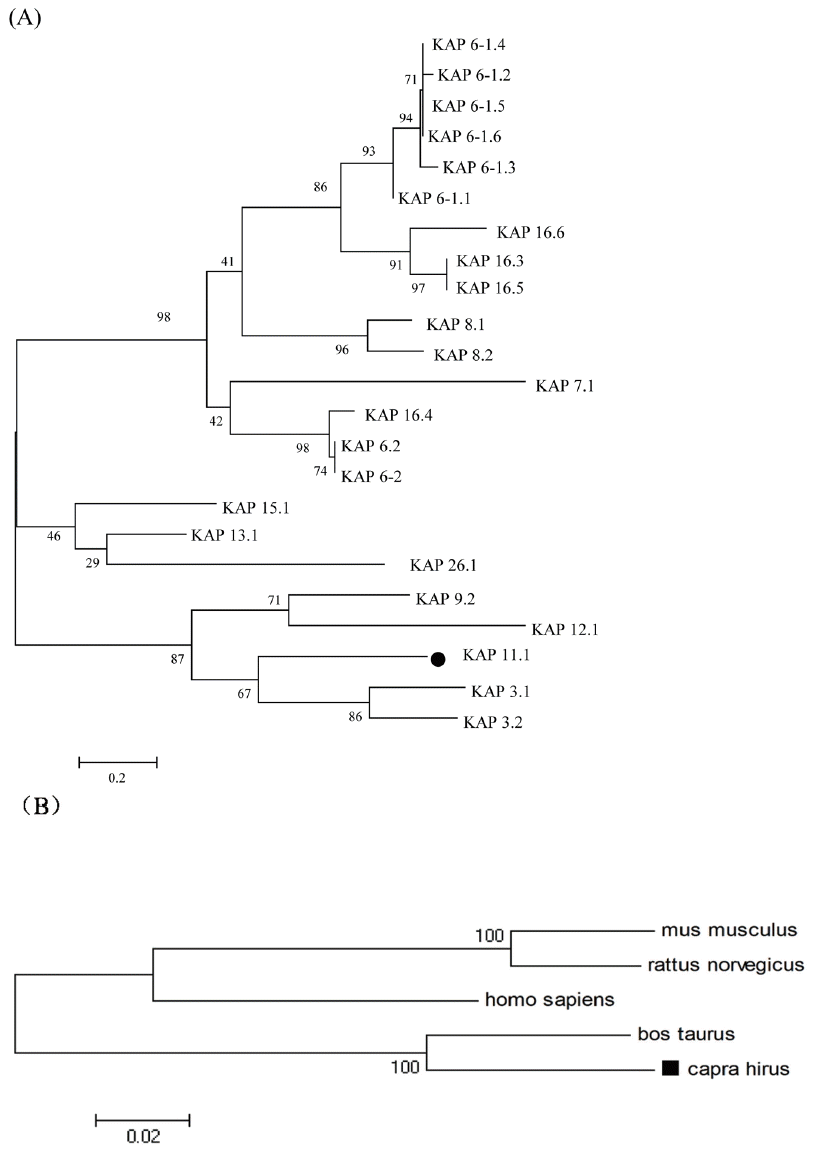
Phylogenetic tree of KAP amino acid sequences. (A) Phylogenetic tree for 23 KAP amino acid sequences from goat or sheep. (B) Phylogenetic tree for KAP11.1 amino acid sequence in five species. The phylogenetic tree was constructed by the neighbor-joining method using MEGA 4.1 software (http://www.megasoftware.net/). KAP, Keratin-associated protein.
The bioinformatics prediction of protein secondary structure indicated that KAP11.1 had a high proportion of random coil (76.73%) and that the rest of KAP11.1 was extended strand (23.27%). When the sequence was analyzed by ProtScale, KAP11.1 had a maximal hydrophobicity value of about −2 (Figure 4A). The transmembrane domain prediction was performed using TMHMM program (Figure 4B). The result showed that KAP11.1 protein lacked a transmembrane domain, and it was neither the recptor nor located in the membrane. A pubertal mammary gland (PMG) structural domain, putatively involved in KAP11.1 protein function, was found using Pfam. This domain was also found in human and cattle. The deduced signal peptide was identified using SignalP and a subcellular localization prediction was carried out using the EpiLoc program. The results showed that KAP11.1 lacked a signal peptide, and it was most likely located in the cytoplasm (Figure 4C and 4D). This prediction was in accordance with the result of Transmembrane domain prediction. The phosphorylation site prediction was performed by NetPhos program. Eight Ser and nine Thr residues may be phosphorylation sites in the KAP11.1 protein (Figure 4E).
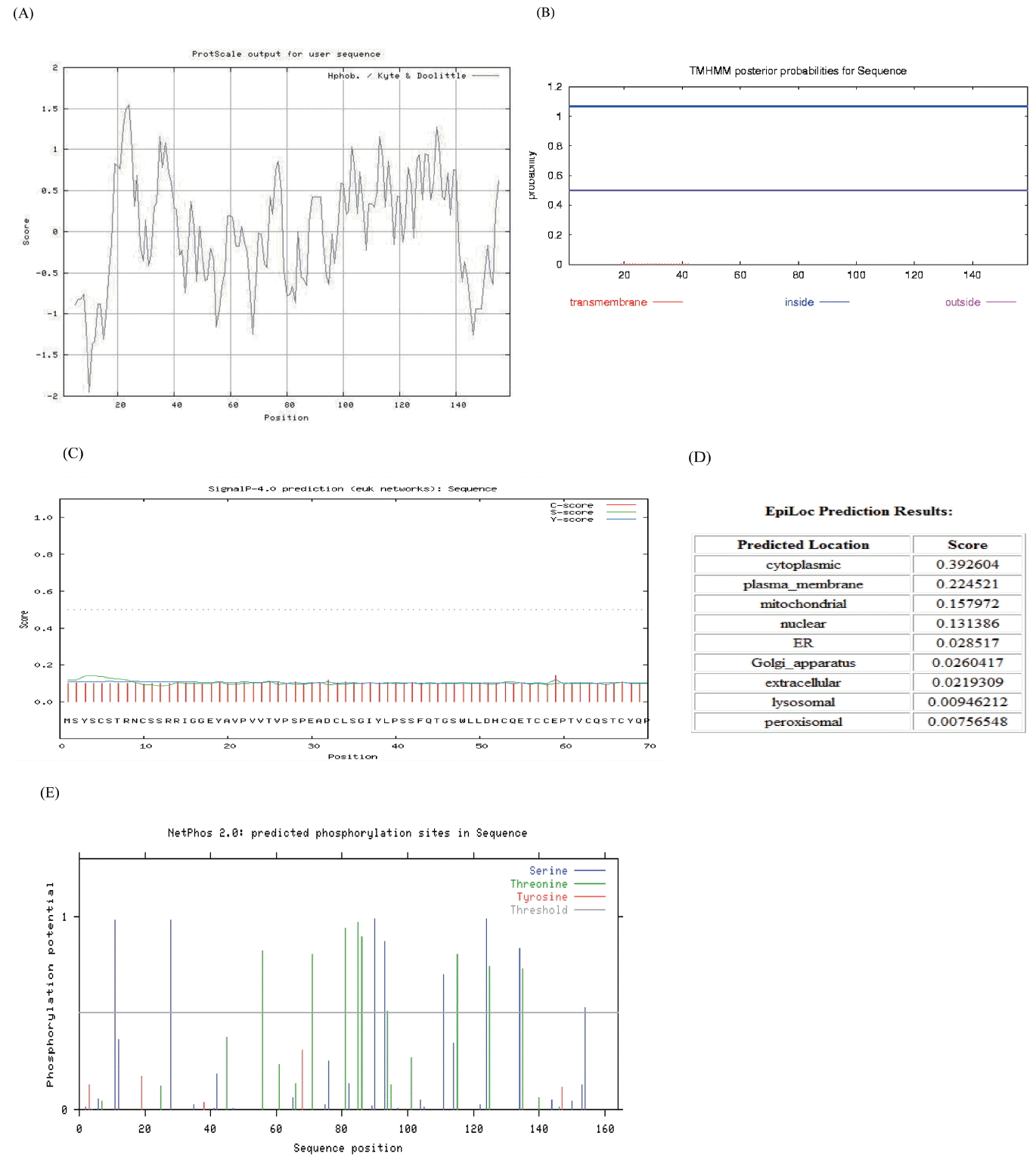
Sequence and bioinforematics analysis of the Liaoning cashmere goat KAP11.1. (A) ProtScale hydrophobicity prediction for the Liaoning cashmere goat KAP11.1 protein. Abscissa is the sequence site and the ordinate is the amino acid scale value. In Kyte & Doolittle amino acid scores, a value >0 represents hydrophobicity, and a value <0 represents hydrophilicity. (B) TMHMM analysis (http://www.cbs.dtu.dk/services/tmhmm/) and forecast of the transmembrane domain of the Liaoning cashmere goat KAP11.1 protein. Red line is transmembrane, blue line is inside, and pink line is outside. (C) The signal peptide analysis of the Liaoning cashmere goat KAP11.1 protein. (D) The subcellular location predictions for the Liaoning cashmere goat KAP11.1 protein. (E) The phosphorylation site analysis of the Liaoning cashmere goat KAP11.1 protein. KAP, Keratin-associated protein.
Tissue distribution of the Liaoning cashmere goat KAP11.1 mRNA
To determine the steady-state level expression of the KAP11.1 gene in different goat tissues, semi-quantitative RT-PCR analysis was carried out with total RNA extracted from skin, heart, liver, spleen, lung and kidney (Figure 5). The KAP11.1 gene was expressed in skin, heart and liver, but not expressed in spleen, lung and kidney.
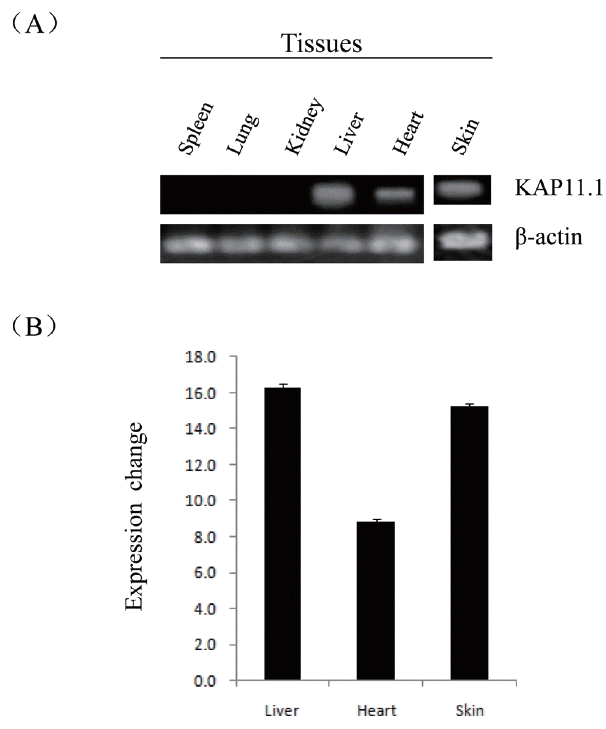
Expression of KAP11.1 gene in different tissues. (A) Semi-quantitative RT-PCR detection of KAP11.1 transcripts while β-actin gene as a loading control. (B) The band using gel was analyzed by BandScan software version 5.0. KAP, Keratin-associated protein; RT-PCR, (Reverse transcription-polymerase chain reaction).
Expression of the Liaoning cashmere goat KAP11.1 mRNA in primary hair follicles and secondary hair follicles
Real time PCR showed that the reference gene (β-actin) and the target gene (KAP11.1) obtained preferable amplification and melting curves. The amplification efficiency was high, and the melting curves were all single apex, indicating good specificity.
The levels of KAP11.1 gene expression in the primary and secondary hair follicles during anagen and catagen were different. In primary hair follicles, the expression of KAP11.1 was higher in catagen than in anagen. In the secondary hair follicles, the expression of KAP11.1 was higher in anagen than in catagen (Figure 6). In telogen, we could not separate the primary and secondary hair follicles because of hair follicle degradation.
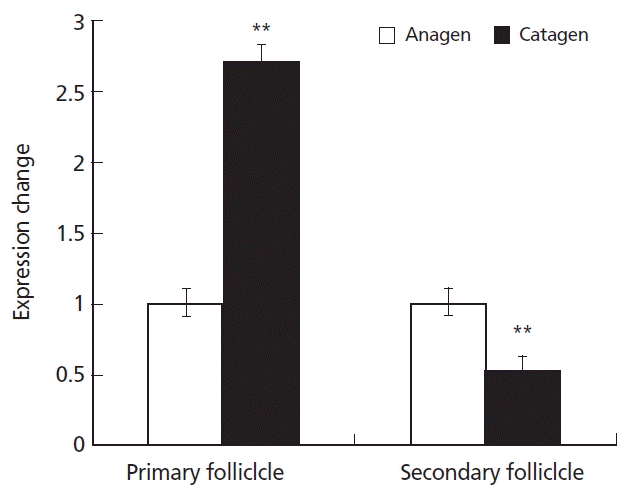
Expression of KAP11.1 gene in the primary hair follicles and secondary hair follicles. Data are expressed as mean±standard deviation of three independent experiments and ** p<0.01 indicates statistically significant expression of KAP1.1 gene in catagen compared with in anagen. KAP, Keratin-associated protein.
Expression location of the Liaoning cashmere goat KAP11.1 mRNA in primary hair follicles and secondary hair follicles
The positive results of in situ hybridization (ISH) was seen based on the observation by Microscope, that was grey-green hybridization signal, ISH results showed that KAP11.1 had a strong signal in the inner root sheath of primary and secondary hair follicles. Additionally, a clear signal in matrix cell were found as well, but no signal in the medulla layer (Figure 7). Each experiment had a separate control, in which no expression signal was found.
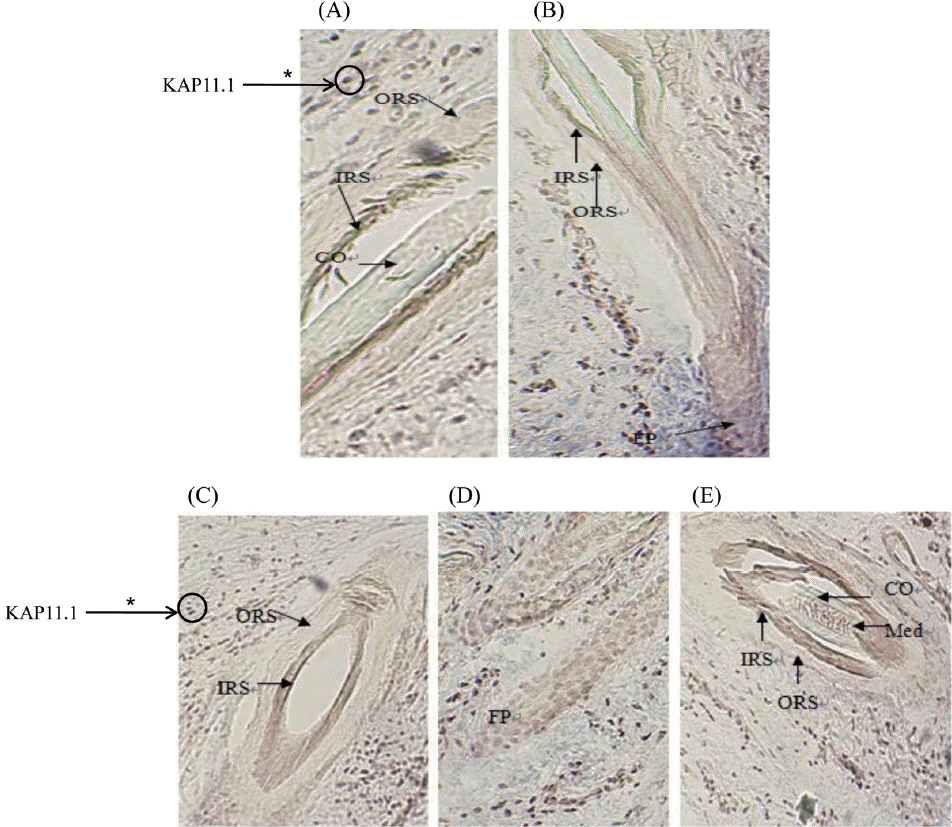
Expression location of KAP11.1 gene in primary hair follicles and secondary hair follicles. (A) The expression of KAP11.1 gene in the primary hair follicle (slitting section, 100×). (B) The expression of KAP11.1 gene in the secondary hair follicle (slitting section, 100×). (C) The expression of KAP11.1 gene in the secondary hair follicle (transverse line, 100×). (D) The expression of KAP11.1 gene in the follicle papilla (slitting section, 100×). (E) The expression of KAP11.1 gene in the primary hair follicle (transverse line, 100×). The areas of KAP11.1 gene expression in the image were marked with asterisk (*). KAP, Keratin-associated protein; ORS, outside root sheath; IRS, inner root sheath; CO, cortical layer; MED, medulla layer; FP, follicle papilla.
DISCUSSION
Liaoning cashmere goats are high-yield cashmere goats native to Liaodong Peninsula. This cashmere goat has a particular feature, which is its fine variable cashmere fleece. Thus it is termed the Liaoning cashmere goat [10]. Cashmere has a very good textile performance, which is closely related to the keratin component and structure. Therefore, clarifying the protein composition of cashmere fibers has both an important theoretical and practical significance.
In the last decades, significant advances have been made toward the characterization of human, goat and ovine KAPs family [11–14]. However, studies on KAP11.1 gene are few. In a previous study, a novel KAP gene of Liaoning cashmere goat, KAP11.1 was isolated. An assumed TATAAbox was located in almost all of the KAP gene coding strands 100 bp upstream of the initiation codon. In this study, we did not find the TATAAbox upstream from the initiation codon [15].
A bioinformatics analysis can predict the structure of a gene and protein, thus laying a foundation for further studies of protein function [16]. Based on the amino acid composition, KAP 11.1 belongs to the high/ultrahigh cysteine (HS) KAPs. KAPs play an important part in regulating cashmere quality, which may be caused by the interaction of KAP–KAP and KIF–KAP [17,18] The data indicated that microfilament cytokeratin (IFs) genes were expressed first, followed by the expression of high glycine/tyrosine KAPs genes (including KAP6, KAP7, KAP8), and finally the high sulfur KAPs genes were expressed [19]. HS KAPs are essential for the formation of hair shafts through their extensive disulfide bond cross-linking to the abundant cysteine residues of hair keratins or hydrophobic interactions with keratins [20–23]. Compared with other KAPs of goat or sheep, KAP11.1 had a high arginine content, which was similar to KAP6.2, and a high content of glutamine, isoleucine and threonine, which were the same as KAP1.3. The content of methionine, phenylalanine and tryptophan were also similar to KAP1.3. The results above can also be demonstrated through an evolutionary tree (Figure 3).
The PMG protein family consists of anagen-specific protein KAPs (PMG1 and PMG2). Both genes are expressed in the growing hair follicles in skin as well as in sebaceous and eccrine sweat glands. Interestingly, expression is also detected in the mammary epithelium where it is limited to the onset of the pubertal growth phase and is independent of ovarian hormones. Their broad, developmentally controlled expression pattern, together with their unique amino acid composition, demonstrate that PMG1 and PMG2 constitute a novel KAP gene family participating in the differentiation of all epithelial cells forming the epidermal appendages [24]. In this study, a PMG structural domain putatively involved in KAP11.1 protein function, which was also found in KAP3.1, KAP12.1, KAP13.1, KAP15.1, and KAP26.1. KAP11.1 showed high homology with KAP3.1 and KAP12.1. However, a low homology was shown with KAP13.1, KAP15.1, and KAP26.1. These results can also be demonstrated through an evolutionary tree (Figure 3).
There may be many phosphorylation sites in the KAP11.1 protein. Scientists have confirmed thatphosphorylation plays an important role in regulating the formation of keratin filament and cell cycle [25].
The results of semi-quantitative RT-PCR in adult goat tissues indicated that KAP11.1 mRNA was strongly expressed in skin, heart and liver while its expression was restricted in lung, spleen, and kidney in our study. Moreover, there has been no investigation of expression in KAP11.1 in any species tissues. But, Rogers et al demonstrated that a large number of KAP family members were only expressed in hair follicles [26]. These results may suggest that KAP11.1 played a more important role in skin, heart and liver than in low expression level tissues. So the function of KAP11.1 in the skin, heart and liver required further study.
Generally, a cashmere goat has heterogeneous fleece composed of cashmere and wool that are produced by the secondary and the primary hair follicles, respectively. The average diameter of coarse wool is about 80 μm and its economic value is not high. The fineness of villus, which is a high textile raw materials, is less than 18 μm um. We used real time PCR to detect the levels of KAP11.1 gene expression in the primary and secondary hair follicles during anagen, catagen and telogen. The results showed that KAP11.1 has a higher expression in catagen than in anagen in the primary hair follicles. However, in the secondary hair follicles, KAP11.1 has a signigicantly higher expression in anagen than in catagen. In telogen, we could not separate the primary and secondary hair follicles because of hair follicle degradation. The expression of the KAP11.1 gene was similar to β-actin in skin tissue. Therefore, we conclude that the KAP11.1 gene has a function in regulating cashmere fineness.
This study, ISH was used to detect KAP11.1 gene expression location. ISH results showed that KAP11.1 had a strong signal in the inner root sheath of primary and secondary hair follicles. All these indicate that KAP11.1 is the structural components of inner root sheath, participating in the formation of inner root sheath of villi, playing an important role in the cashmere growth and quality. But no signal in the outer root sheath (ORS), sebaceous glands and medulla layer, explained that KAP11.1 is not involved in the composition of ORS, sebaceous gland and hair medulla. Besides, a clear signal in matrix cell were found in Liaoning cashmere goat. This may be one of the reasons for different cashmere quality in the different species.
CONCLUSION
We analyzed the protein features and gene expression of KAP11.1 by bioinformatics, and we forecast its protein structure and function. This is a solid foundation for further studying the function of the KAP11.1 gene in regulating hair follicle growth. We analyzed the expression of the KAP11.1 gene in Liaoning cashmere goats by RT-PCR and real-time PCR methods. This provides a molecular biological basis for further understanding the function of genes involved in regulating the diameter of fibers.
ACKNOWLEDGMENTS
Distinguished professor support plan of Liaoning province, Scientific Research Fund of Liaoning Provincial Education Department (No.L2014425), National Natural Science Foundation of China (No. 31172188).
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
