Rapid Genotyping of MSTN Gene Polymorphism Using High-resolution Melting for Association Study in Rabbits
Article information
Abstract
The myostatin (MSTN) gene, as a negative regulator of skeletal muscle growth, has been proposed to be associated with production traits in farm animals. In the present study, a T/C variant at −125 bp (relative to ATG start codon) of 5’regulatory region of rabbit MSTN was identified by direct sequencing. Two hundred and twenty two rabbits, which were randomly sampled from 3 breeds (Ira rabbits, Champagne rabbits and Tianfu black rabbits), were genotyped by high-resolution melting (HRM). Comparing the genotyping results of 47 samples with direct sequencing, the HRM showed high sensitivity (0.96) and high specificity (0.98). In the three rabbit breeds, the allele C was the predominant allele. The polymorphic site showed high heterozygosity (He = 0.48) and high effective number of alleles (Ne = 1.91). The genetic diversity was reasonably informative (0.25<PIC<0.50). The association analysis showed that the genotype TC had significant effect on the 84-d-weight of rabbits compared with genotype CC (p = 0.047). In contrast, the genotypes had no significant effect on other production traits. These results showed that HRM could be effectively used for genotyping analysis of MSTN gene. The T/C variant in 5’regulatory region of MSTN might be one of the candidate SNP loci affecting the trait of 84-d-weight.
INTRODUCTION
The myostatin (MSTN) gene is known as GDF8 (growth differentiation factor 8) belonging to TGF-β (transforming growth factor-beta) family, which is also considered as a negative regulator of skeletal muscle growth. It has been proved that the MSTN knockout mice have significantly larger body weight and more widespread increase in skeletal muscle mass than that of the wild-type (McPherron et al., 1997). The polymorphisms of this gene have been reported to be associated with production traits which make it a candidate gene to enhance the productivity of livestock and fowl (Lu et al., 2011; Han et al., 2012; Zhang et al., 2012). Rabbits have traits of economic importance like high rate of reproduction, early maturity, high growth rate and efficient feed utilization (Bindu et al., 2012). In previous studies, the rabbit MSTN was considered as one of the candidate genes of meat production traits and its single nucleotide polymorphisms (SNP) were only focused on the exon, intron and 3’-untranslated region (Fontanesi et al., 2008; Fontanesi et al., 2011; Kurkute et al., 2011). The mutation in 5’regulatory region of MSTN could lead to changes of the gene expression and thereby influence growth and carcass traits. However, the research of 5’regulatory region of rabbit MSTN is limited.
Direct sequencing technique is considered as the gold-standard approach for genotyping analysis and expected to have almost 100% sensitivity (Laurie et al., 2009). Many techniques have been developed to reduce the use of direct sequencing and control costs while maintaining sensitivity and specificity. The sensitivity of early methods, such as single-stranded conformation polymorphism analysis (SSCP) (Kunhareang et al., 2009) and denaturing gradient gel electrophoresis (DGGE) (Kirk et al., 2002), are low (70% to 80%) and require labor intensive and considerable technical skills. Denaturing high performance liquid chromatography (DHPLC) has high sensitivity (>96%) if amplicons are well-designed and analysis temperatures are optimized (Bunn et al., 2002).
High-resolution melting (HRM) analysis is a well established, closed tube, rapid, and cost-effective SNP genotyping technique based on the analysis of the melting profile of PCR products, using intercalating fluorescent dyes to monitor the transition during heating (Martino et al., 2010). As the temperature increases, the fluorescence decreases, producing a characteristic melting profile (Montgomery et al., 2007). The presence of a single nucleotide mutation in the amplicon will change the melting temperature (Tm) and the shape of the melting curve.
MSTN gene and its genetic polymorphism have been widely proposed to significantly associate with economic traits in farm animals. Therefore, developing a simple and economical genotyping method for MSTN gene is necessary for marker-assisted selection and breeding. HRM, characterized by high genotyping sensitivity and specificity, is being employed for MSTN genotyping in rabbit. In this study, direct sequencing was used to identify single nucleotide polymorphisms in the 5’regulatory region of rabbit MSTN. Then, genotyping of 222 randomly sampled individuals from the Ira rabbits, Champagne rabbits and Tianfu black rabbits was performed by HRM. We first compared the genotyping results of 47 samples with direct sequencing. Meanwhile, the association between MSTN polymorphism and rabbit production traits was subsequently analyzed by least square method.
MATERIAL AND METHODS
Animals, growth traits and carcass traits collection
This study was conducted in a total of 222 commercial meat rabbits including 88 Ira rabbits, 86 Champagne rabbits and 48 Tianfu black rabbits. The nutritional levels and feeding management were described previously (Zhang et al., 2011), with CP = 16%, DE = 10.8 MJ/kg. The carcasses were dissected according to the norms of the World Rabbit Science Association, and the data of growth traits and carcass traits were collected. The growth traits included 35-d-weight, 70-d-weight, and 84-d-weight; the carcass traits included eviscerated weight, semi-eviscerated weight, eviscerated slaughter rate, and semi-eviscerated slaughter rate (Blasco et al., 1993).
Genomic DNA isolation and mutation screening
Ear tissues were collected for genomic DNA extraction using AxyPrep Genomic DNA Miniprep Kit (Axygen, USA) and the DNA was stored at −20°C. A polymerase chain reaction (PCR) primer pair (Forward 5’-GAC TTGTGACAGACAGGGTTT-3’ and Reverse 5’-AATCT TTATGGCTTCTATTCTTG-3’) was newly designed in this study to amplify the 5’regulatory region according to the reference sequence of rabbit MSTN (GenBank accession number GU938461.1) by Primer 5.0, the PCR product size was 437 bp. The PCR reaction was performed with the following condition: one denaturation cycle at 94°C for 5 min; 35 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 40 s; and ended with an extension cycle at 72°C for 10 min. The 30 μl reaction volume included 15 μl 2×Taq PCR MasterMix (TIANGEN, Beijing, China), 3 μl DNA template (20 ng/μl), 9.6 μl ddH2O, 1.2 μl of each primer (10 pmol/μl). PCR products from 16 rabbits were randomly selected to scan the genetic polymorphism in the 5’regulatory region using direct sequencing method by Shanghai Invitrogen Biotechnology Co., Ltd (Shanghai, P. R. China).
Genotyping using HRM
A primer pair (Forward 5’-TCATAGACCCTGACGAC ACT-3’ and Reverse 5’-CTTTTCAGTAATGGCAAGC-3’) was newly designed to amplify the small fragment containing the SNP revealed by direct sequencing, and the product size was 141 bp. The 10 μl reaction volume included 5 μl 1×SsoFast EvaGreen supermix (Bio-Rad, USA), 1 μl DNA template (20 ng/μl), 3.2 μl ddH2O, 0.4 μl of each primer (10 pmol/μl), and was pipetted into each well on the 96-well PCR plate. The cycling condition was: one denaturation cycle at 98°C for 3 min; 40 cycles of 5 s at 98°C and 25 s at 57°C; following the step of 95°C for 10 s; a melting step ramped from 65°C to 95°C rising by 0.2°C each step with date acquisition for 10 s. All samples were amplified in duplicate, each plate included DNA from sequencing-verified wild-type, heterozygote, homozygote and no template control (NTC). The PCR reaction was performed on Bio-Rad CFX96 real-time PCR detection system (Bio-Rad, Inc., Hercules, CA, USA). The amplicon was subsequently subjected to HRM analysis (Martino et al., 2010).
Data analysis
The DNA sequences were assembled and analyzed using the DNAstar program (DNAS Inc, Madison, WI, USA). HRM curve data were analyzed using the Precision Melt Analysis™ software (Martino et al., 2010). Allele frequencies, genotype frequencies, heterozygosity (He), effective number of alleles (Ne) and Polymorphic Information Content (PIC) were estimated based on the study of previously described (Bindu et al., 2012). The effects of genotype on the traits were analyzed by the general linear model (GLM) procedure of SAS 9.2 program according to the following statistical model:
Where Yijk is studied traits, μ is the overall mean of observations, Gi is the fixed genotype effect of the MSTN mutation site, Sj is gender effect, Bk is breed effect, and eijk is the residual error.
RESULTS AND DISCUSSION
Identification of SNP in rabbit MSTN
In previous studies, SNPs in rabbit MSTN was focused on the exon, intron and 3’-untranslated region. Fontanesi et al. (2008) sequenced the entire MSTN in four different breeds of rabbit and only detected one SNP in intron 2 (c.747+34C>T). Then Fontanesi et al. (2011) re-sequenced the MSTN including the three coding exons in 14 rabbits with different conformations and muscle mass, and identified another three SNPs of MSTN in exon 1 (c.108C>T), exon 2 (c.713T>A) and 3’-untranslated region (c.*194A>G). Kurkute et al. (2011) detected all three exons of MSTN in three rabbit breeds and found two mutations in exon 1 (c.1A>G) and exon 3 (c.799G>A).
In the present study, a T/C variant at −125 bp (relative to ATG start codon) of rabbit MSTN was identified by direct sequencing method (Figure 1). The T/C variant is a newly discovered variant in 5’regulatory region of MSTN. In silico analysis revealed that the allele C increased a transcription factor binding site of AP-2 and decreased a transcription factor binding site of CAAT (http://tfbind.hgc.jp/). The CAAT box is a motif present in the conserved sequence upstream of the start points in eukaryotic transcription units, which is recognized by a large group of transcription factors. AP-2 transcription factors are localized predominantly in the nucleus where they bind to target sequences and regulate transcription of target genes (Dawid et al., 2005). The change of these transcription factor binding sites might affect RNA transcription, which should be further investigated.
Genotyping of SNP in rabbit MSTN by HRM
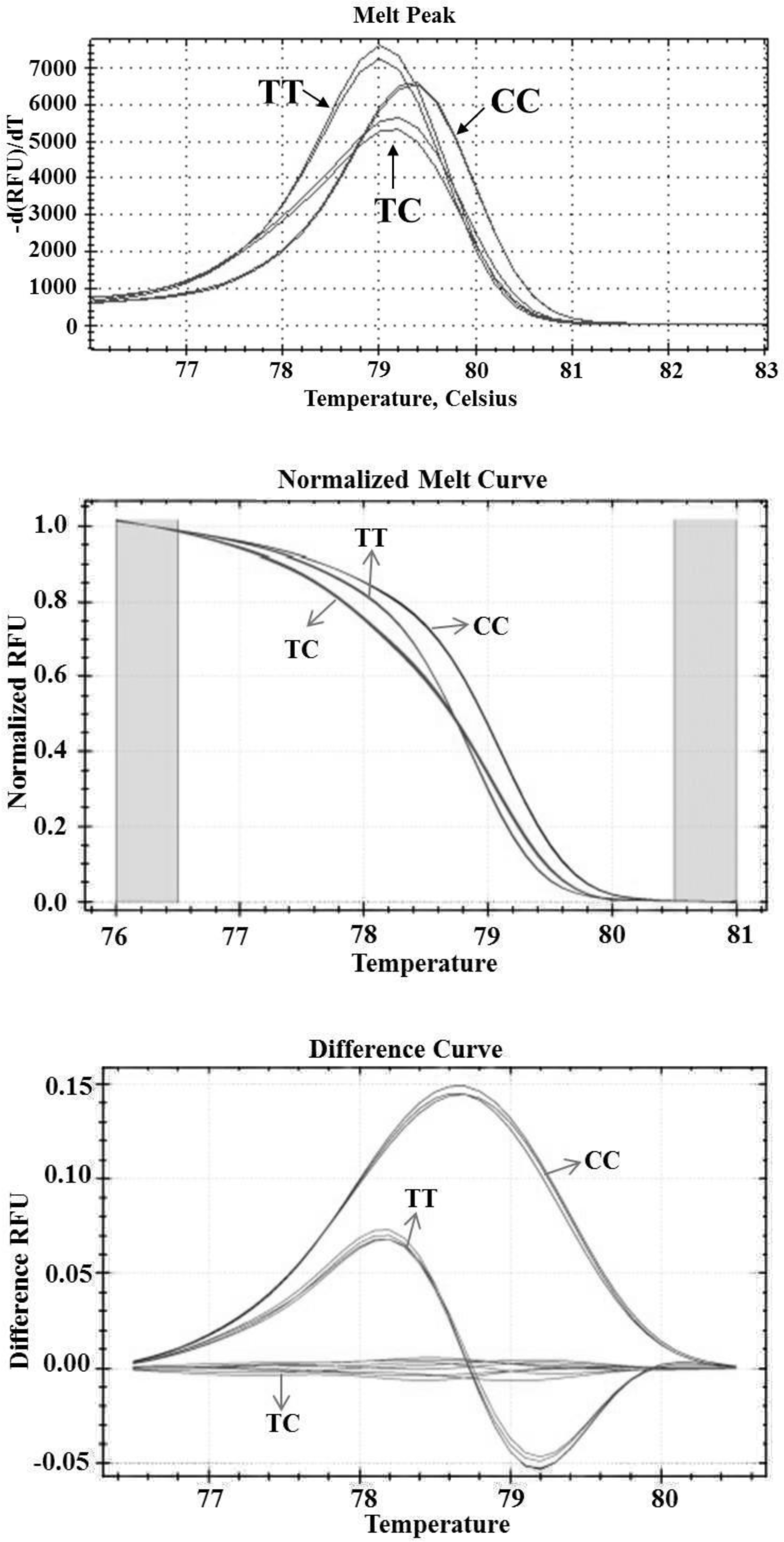
HRM was performed on sequencing-verified samples in order to determine the Tm and characterize of the melting curve profiles of different genotypes. In this study, each genotype produced unique melt curves (amplitude and/or shape of the curves) and could be discriminated directly (Figure 2). The genotyping of SNP in MSTN by HRM was done as following: first, the curves were auto grouped by the software based on the melt transition, and then the observer confirmed the difference between Tm and melt curves. There was no difference in the classification by the observer compared to that grouped by the software.
The homozygous CC (79.4°C) melted at a higher temperature than the homozygous TT (79.0°C) due to the higher stability of the G ≡ C pairing compared to A = T. The difference in temperature could unambiguously differentiate the two homozygous sequences. The heterozygotes were identified based on the changes in the shapes of the melt curves as compared to homozygotes. The presence of both forms of homozygotes together with heteroduplexes results in a change in the melting pattern characterized by a flatter curve shape/melting transition (Raghavendra et al., 2011), as observed in the normalized melting curves (Figure 2).
Validation of sensitivity and specificity of genotyping by HRM
We selected 47 samples which were on the same plate to be sequenced for verifying the sensitivity and specificity of genotyping by HRM. Sequencing was performed by Shanghai Invitrogen Biotechnology Co., Ltd (Shanghai, P. R. China). The sensitivity and specificity of HRM were 0.96 (45 true positive/(45 true positive+2 false negative)) and 0.98 (92 true negative/(92 true negative+2 false positive)), respectively. Recent studies have also reported the sensitivity of HRM close to 100% except in the situation where amplicons have a high GC content (Garritano et al., 2009; Laurie et al., 2009).
Direct sequencing is the gold-standard approach for detecting SNPs, but involves additional steps leading to a higher turnaround time. HRM is a closed tube, fast method following PCR amplification and eliminates the possibility of template contamination, where samples can be analyzed in a single run (Raghavendra et al., 2011). HRM does not have 100% sensitivity; however, mutation scanning is a trade-off between the speed, cost, and sensitivity. It is worth to emphasizing how rapid this technique is: a plate of 96 samples can be melted in 10 min (Laurie et al., 2009). SsoFast EvaGreen supermix was used in this study. EvaGreen dye is a fluorescent nucleic acid dye with spectral properties similar to SYBR Green I and fluorescein. Unlike SYBR Green I, EvaGreen dye exhibits very low PCR inhibition. Thus, it can be used at high concentrations to generate greater fluorescent signals and provide increased sensitivity. Consequently, HRM is a better candidate to detect sequence mutations on a large scale, as it is much quicker than direct sequencing.
Genotype and allele frequencies in 5’ regulatory region of rabbit MSTN
The genotype and allele frequencies in the 5’ regulatory region of MSTN in different rabbit breeds is shown in Table 1. Allele C was the predominant allele with frequencies of 0.56 in Iraq, 0.58 in Champagne and 0.76 in Tianfu rabbits, respectively. The frequencies of genotype TC for Iraq and Champagne rabbits were the highest (0.64 and 0.59, respectively), but the frequency of genotype CC was the highest in Tianfu rabbits (0.56). The polymorphic site showed high heterozygosity (He = 0.48) and a high effective number of alleles (Ne = 1.91), and the genetic diversity was reasonably informative (0.25<PIC<0.50) in the three rabbits breeds. The genetic diversity revealed higher genetic variation and a selective potential that could be expected to get more genetic progress. In previous studies, a C-T transition at position 34 of MSTN intron 2 has been reported in New Zealand White, Soviet Chinchilla, Belgian Hare and several other rabbit breeds (Fontanesi et al., 2008; Bindu et al., 2012). In the present study, we randomly selected three rabbit breeds that were different from previous studies. The results showed similar frequencies and genetic characteristics.
Association between MSTN genotypes and production traits in rabbits
Some studies have showed that mutations in MSTN can inactivate its expression and further affect muscle fiber development and cause dramatic muscularity and a “double muscling” phenomenon in cattle (Wiener et al., 2002; Esmailizadeh et al., 2008; Grisolia et al., 2009). In the present study, the association analysis (Table 2) showed that different genotypes had a significant effect on the 84-d-weight of rabbits, individuals with the genotype TC indicated a significant association with higher 84-d-weight than genotype CC (p = 0.047). But MSTN genotypes had no significant effect on other production traits. Similar findings have been reported previously. Fontanesi et al. (2011) indicated that MSTN is not an important source of variability for performance traits, at least in the rabbit population analyzed. Bindu et al. (2012) also showed that a single nucleotide polymorphism (C-T transition) at position 34 of intron 2 of rabbit MSTN was associated with higher body weight but the difference in body weight among different genetic groups was not statistically significant.
As previously mentioned, the T/C variant at −125 bp caused the change of transcription factor binding sites which might affect RNA transcription. Sadkowski et al. (2008) also showed that polymorphism in the 5’-flanking region of MSTN was associated with the level of mRNA in skeletal muscle of bulls. Lin et al. (2002) showed that MSTN expression levels of Asian pigs increased markedly at 3 moths and maintained a constant level thereafter. Sun et al. (2012) also demonstrated that MSTN expression levels in muscle of Hu sheep were lowest at two days old, and tended to increase with the increasing age before 60 days. For these reasons, we could assume that MSTN expression levels might be relatively low in the initial growth process. Therefore, the T/C variant at −125 bp of rabbit MSTN had no significant effect on 35-d-weight and 70-d-weight. As the rabbit grows, the expression levels should gradually increase and the inhibitory effect on muscle growth should also gradually become obvious. At 84-d, the rabbit growth reached a relatively stable period and the inhibitory effect on muscle growth of MSTN might be most obvious. That caused the individuals with the genotype TC to have a significantly higher 84-d-weight than genotype CC (p = 0.047).
CONCLUSION
These results showed that HRM could be effectively used for genotyping analysis of the MSTN gene. The T/C variant in 5’regulatory region of MSTN might be one of the candidate SNP locus affecting the trait of 84-d-weight.