Naturally Occurring Lactic Acid Bacteria Isolated from Tomato Pomace Silage
Article information
Abstract
Silage making has become a significant method of forage conservation worldwide. To determine how tomato pomace (TP) may be used effectively as animal feed, it was ensilaged for 90 days and microbiology counts, fermentation characteristics and chemical composition of tomato pomace silage (TPS) were evaluated at the 30th, 60th, and 90th days, respectively. In addition, 103 lactic acid bacteria were isolated from TPS. Based on the phenotypic and chemotaxonomic characteristics, 16S rDNA sequence and carbohydrate fermentation tests, the isolates were identified as 17 species namely: Lactobacillus coryniformis subsp. torquens (0.97%), Lactobacillus pontis (0.97%), Lactobacillus hilgardii (0.97%), Lactobacillus pantheris (0.97%), Lactobacillus amylovorus (1.9%), Lactobacillus panis (1.9%), Lactobacillus vaginalis (1.9%), Lactobacillus rapi (1.9%), Lactobacillus buchneri (2.9%), Lactobacillus parafarraginis (2.9%), Lactobacillus helveticus (3.9%), Lactobacillus camelliae (3.9%), Lactobacillus fermentum (5.8%), Lactobacillus manihotivorans (6.8%), Lactobacillus plantarum (10.7%), Lactobacillus harbinensis (16.5%) and Lactobacillus paracasei subsp. paracasei (35.0%). This study has shown that TP can be well preserved for 90 days by ensilaging and that TPS is not only rich in essential nutrients, but that physiological and biochemical properties of the isolates could provide a platform for future design of lactic acid bacteria (LAB) inoculants aimed at improving the fermentation quality of silage.
INTRODUCTION
Using byproducts of food processing as animal feed has proven economically viable, not only because it is a method of waste and residue disposal but also because it is a real alternative for feeding livestock (Parvin et al., 2010). In China, it is increasingly important to produce highly digestible forage to support the expanding dairy industry. The number of dairy cattle reached 12.6 million in 2010 (Wang et al., 2013). Usually, food processing byproducts contain high amounts of protein and fiber, rendering them suitable for animal feed production (Council for Science and Technology, 2005). Tomato pomace (TP), a byproduct of the tomato juice industry, contains an abundance of nutrients, such as vitamins, minerals, vegetable fiber and protein (Delvalle et al., 2006; Abdollahzadehi et al., 2010). TP is produced in vast amounts annually in the Bayannur region, the largest tomato production base in Inner Mongolia, generating 1.8 to 2.3 million tons of tomato and 100,000 tons of tomato byproducts. The high water content (75%) of this byproduct limits its length of storage. Thus, TP is often dried. Dried TP is fed to dairy cows and sheep (Belibasakis and Ambatzidiz, 1995; Zheng et al., 2011a), but artificial drying increases the price of TP substantially; thus, much of the pomace that is produced is discarded (Weiss et al., 1997; Nemat Ziaei and Sadrollah Molaei, 2010). In the light of the above, it is obvious that TP has a high potential to become a highly nutritious and economic livestock feed if it can be economically preserved.
TP can be fed to ruminant animals for longer periods of time without spoilage, when it is ensiled with or without additives (Denek and Can, 2006). Of the many factors that influence this process, the number and type of microorganisms that dominate the fermentation process often dictate the final quality of the silage (Cai, 1999a; Weinberg et al., 2004). The preservation of silage depends on the production of sufficient organic acid mainly produced by lactic acid bacteria (LAB) to inhibit the activity of undesirable microorganisms, such as clostridia and molds, under anaerobic conditions (Cai et al., 1997).
The aim of this study was to determine whether TP could be preserved for a long time as a livestock diet by ensilaging and to examine the LAB in tomato pomace silage (TPS) that are most likely to have significant functions in the fermentation process. In this study, the chemical composition, silage fermentation characteristics and microbiology counts of TPS were measured. Further, LAB isolates from tomato silage were identified based on phenotypic and chemotaxonomic characteristics, 16S rDNA sequence analysis and carbohydrate fermentation tests.
MATERIALS AND METHODS
Tomato pomace sampling and silage preparation
TP was collected from local tomato juice industries (Inner Mongolia, China) on October 7, 2012. TP is a residue that remains after juice is extracted, consisting of remainder flesh, seeds and peels without the addition of bacteria. TPS was prepared in a small-scale fermentation system using a slightly modified method of Tanaka and Ohmomo (1994). Approximately 100 g of tomato pomace, chopped into about 10 mm lengths, was packed into plastic film bags and sealed. The film bag silos were stored outdoors. Three parallel experiments were used for silage treatment. Samples were collected at the 30th, 60th, and 90th days of the ensiling process.
Chemical analysis
TPS and the fresh TP were ground to pass a 1-mm screen and analyzed in 3 parallel experiments. Dry matter (DM), crude protein (CP), and ash content were determined by standard methods (AOAC, 2000) (930.15, 976.05, and 942.05, respectively). Neutral detergent fiber (NDF) was analyzed with thermostable amylase and sodium sulfite and acid detergent fiber (ADF) was analyzed nonsequentially; results were expressed without residual ash (Van Soest et al., 1991). The pH was measured with a glass electrode pH meter (MP230, Mettler Toledo, Greifensee, Switzerland) and ammonia-N was measured by steam distillation of the filtrates (Xu et al., 2007). The organic acid content was measured per Cai et al. (1999b).
Microbiological analysis and lactic acid bacteria isolates
Each of the samples (1 g) were blended with 9 mL sterilized water and serially diluted (10−1 to 10−5) in sterilized water. LAB were incubated at 37°C for 48 h on lactobacilli MRS agar (Huankai Microbial SCI. and Tech, Co., Ltd. Guangdong, China) under anaerobic conditions (anaerobic jar, AnaeroPack-Anaero, Tokyo, Japan) and measured by plate count. Coliform bacteria were incubated at 30°C for 24 h and counted on blue light broth agar (Huankai Microbial SCI. and Tech, Co., Ltd.). Molds and yeast were incubated for 24 h at 30°C and counted on potato dextrose agar (Huankai Microbial SCI. and Tech, Co., Ltd.). Yeast was distinguished from mold and bacteria by colony appearance and cell morphology. Bacilli and aerobic bacteria were distinguished by colony shape and counted on nutrient agar (Huankai Microbial SCI. and Tech, Co., Ltd.), incubated for 24 h at 30°C under aerobic conditions. Colonies were counted as viable numbers of microorganisms in colony-forming units per gram fresh matter (FM). Each LAB colony was purified twice by being streaked on MRS agar. The purified strains were stored at −80°C in nutrient broth (Difco) and dimethyl sulfoxide at a 9:1 ratio for further analysis (Cai et al., 1999d,e).
Morphological, physiological and biochemical tests
Gram-staining and catalase activity were examined after 48 h of incubation on MRS agar. Gram-positive and catalase-negative bacteria were identified as LAB. Morphological characteristics and gas production from glucose were measured as described (Camu et al., 2007). Growth at various temperatures was measured in MRS agar after incubation at 5°C and 10°C for 10 d and at 45°C and 50°C for 7 d. Growth at pH 3.0, 4.0, 5.0, 6.0, 7.0, and 8.0 was monitored in MRS broth after incubation at 37°C for 7 d. Salt tolerance of LAB was tested in MRS agar that containing 3.0% and 6.5% NaCl.
Carbohydrate fermentation tests were performed using API 50 CH strips (BioMérieux, France) for 49 compounds and 1 control per the manufacturer’s instructions; reactions were measured after incubation at 37°C for 48 h. The organisms were identified using API LAB Plus, version 3.3.3 from the BioMérieux and Analytab Products database for comparison of assimilation and fermentation patterns.
PCR amplification, sequencing and analysis of 16S rRNA genes
Cells were grown at 37°C for 8 h in MRS broth and used for DNA extraction and purification (Meroth et al., 2003). The 16S rRNA coding region was amplified by PCR in a thermal cycler (Analytikjena, Flexcycler Block, Germany) using reagents from the Takara Taq PCR Kit (Takara Biotechnology Co., Ltd, Dalian, China).The primers were prokaryotic 16S ribosomal DNA universal primers 27F (5′-GAGTTTGAT CCTGGCTCA-3′) and1492R (5′-TACCTTGTTACGACTT-3′).
The PCR performed was as follows: 50 μL PCR reactions contained 0.25 μL Takara Taq (5 U/μL), 1 μL 27F (10 μmol/L) and 1492R (10 μmol/L) primer, 5 μL 10×PCR buffer (Mg2+-free), 3 μL MgCl2 (25 μmol/L), 4 μL dNTP mixture, 2 μL template DNA and ddH2O up to 50 μL. The PCR program was 94°C for 5 min, 30 cycles at 94°C for 1 min, 53°C for 1 min and 72°C for 2 min and 72°C for 10 min.
16S rRNA gene sequences were compared using GenBank and the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequence information was introduced into CLUSTALW for assembly and alignment (Thompson et al., 1994). The 16S rDNA sequences of the isolated LAB strains were compared with sequences of LAB strains in NCBI (Figure 1). Nucleotide substitution rates (Knuc values) were calculated (Pang et al., 2012) and phylogenetic tree was constructed using the neighbor-joining method (Ennahar et al., 2003). Bacillus subtilis NCDO 1769 was used as an outgroup organism (Duan et al., 2008). The topologies of the tree was evaluated by bootstrap analysis of the sequence data using Molecular Evolutionary Genetics Analysis (MEGA) 5 (Tamura et al., 2007), based on 1,000 random resamplings (Eitan et al., 2006). The sequences were aligned with published sequences of type strains in GenBank.
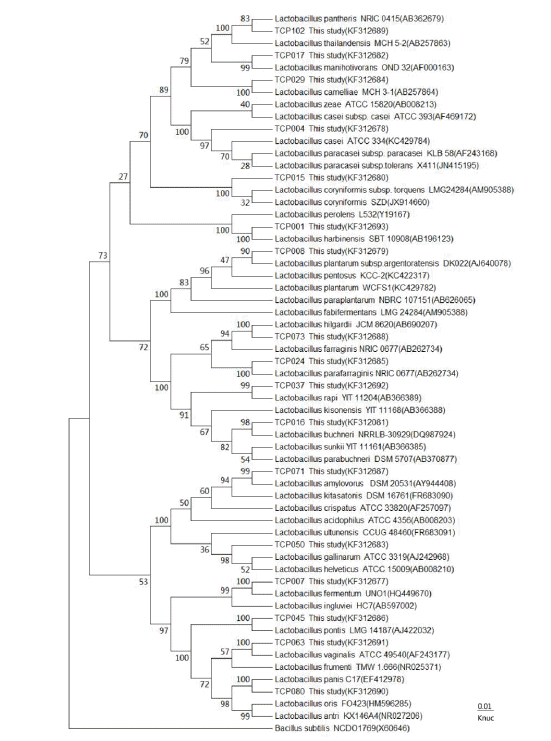
Phylogenetic tree showing the relative positions of the representative isolates isolated from tomato pomace silage as inferred by the neighbor-joining method of complete 16S rDNA sequences. Bootstrap values for 1,000 replicates are shown at the nodes of the tree. Bacillus subtilis is used as an outgroup. The bar indicates 1% sequence divergence. Knuc = Nucleotide substitution rate.
Nucleotide sequence accession number
The nucleotide sequences for the 16S rRNA genes were deposited into GenBank under the following accession numbers: KF312693, KF312677, KF312678, KF312679, KF312680, KF312681, KF312682, KF312683, KF312684, KF312685, KF312686, KF312687, KF312688, KF312689, KF312690, KF312691, and KF312692 for the representative strains TCP001, TCP007, TCP004, TCP008, TCP015, TCP016, TCP017, TCP050, TCP029, TCP024, TCP045, TCP071, TCP073, TCP102, TCP080, TCP063 and TCP037, respectively.
Statistical analyses
Data on chemical composition and silage quality of TPS were subjected to ANOVA and differences between means were assessed by Tukey test using the statistical packages for the social sciences (SPSS 13.0 for Windows; SPSS Inc., Chicago, IL, USA). The effect was considered significant when probability was less than 0.05. All reported values were the average of three replicates.
RESULTS AND DISCUSSION
Microorganism counts, chemical composition and fermentation characteristics of TPS
The counts of viable microorganisms, chemical composition and silage fermentation characteristics of TPS are shown in Table 1. Overall, 107 to 108 (cfu/g FM) LAB, 102 molds, 103 to 105 aerobic bacteria, 102 yeasts and 105 bacilli were found in TPS samples. It is notable that coliform bacteria were not present and that LAB was the dominant group among the microorganisms in the TPS.
Due to the anaerobic fermentation process, the number of aerobic bacteria in the TPS was less than that of fresh TP and continued to reduce with time during the fermentation process. The populations of LAB in TPS (107 to 108 cfu/g FM) increased significantly compared to that of fresh TP (103 cfu/g FM) and kept increasing in the ensiling process, which was good for the conversation of TPS. This great increase was mainly related with the anaerobic environment. 104 coliform bacteria were found in fresh TP, while in the TPS they were totally inhibited. Although the count of molds in the TPS increased compared with that of fresh TP, the number of moulds was still low. Moreover, the count of molds kept reducing with time. The inhibition of undesirable microorganisms, such as coliform bacteria and molds was attributable to the increase of LAB which produce lactic acid that inhibits the growth of these undesirable microorganisms. From a microbiology point of view, therefore, ensilaging TP in this study has been successful as it witnessed a simultaneous time-dependent increase and decrease in desirable and undesirable microbial populations respectively.
The levels of CP, lactic acid, acetic acid, phosphorus, calcium and ammonia-N increased compared with those of the fresh TP and continued to significantly increase during the whole storage period (p<0.05). The ADF content and pH of TPS declined (p<0.05) during storage. The levels of ash, organic matter, ether extract and DM in the TPS did not have significant change (p>0.05) over time. The level of NDF and butyric acid did not change significantly before 60 days (p>0.05) but declined afterwards (p<0.05). Lactic acid ensures long-stem storage of silage by creating a low pH environment to inhibit the survival of undesirable microbes. Phosphorus and calcium are needed for good nutritional value of silage. The decrease of ADF and NDF was attributed to the degradation of fibrous material and this indicated that the indigestible fibrous material hydrolyzed to cause more energy value of silage. Lower ADF and NDF are good for digestion and metabolism in ruminants. In addition, too much ADF and NDF would reduce the dry matter intake of ruminants. It was observed that the color of TP was virtually preserved in the TPS during the ensiling period. For instance the red color of the TP was still maintained in the TPS at the 30th day while its light red and reddish yellow colors at the 60th and 90th days respectively were only slightly and thus inconsequentially, different from the original red color of the TP. The original color of the TP was therefore largely preserved in the TPS. Given the chemical composition and fermentation characteristics of TPS, therefore, ensilaging TP to preserve it would be an economic way of producing a highly nutritive livestock forage alternative.
Morphological, physiological and biochemical properties
The morphology, characteristics and API 50 CH fermentation patterns of representative strains from TPS are shown in Table 2 and 3. A total of 103 (TCP001 to TCP103) LAB strains were isolated from silage, on the basis of their gram-positive reaction, negative catalase reaction and production of lactic acid as the chief fermentation product. Ultimately, they were identified as 17 species and all belonged to genus Lactobacillus, in contrast to previous research. For example, Cai et al. (1998; 1999a,b) noted that the predominant LAB in many forage crops and grasses were lactic acid-producing cocci and that the least frequent were lactobacilli (primarily homofermentative). In our study, the LAB strains were isolated from tomato pomace ensiled at least 30 days, whereas in the research of Cai et al. (1998; 1999a,b), the strains were isolated from fresh materials. This difference may be the one of the reasons for the contrasting results. According to the literature, in the early fermentation, lactic acid-producing cocci reproduced massively and pH fell rapidly. During fermentation, lactic acid-producing cocci were replaced by lactobacilli gradually until lactobacilli became dominant. In addition, the distinct materials in tomatos may also contributed the difference in LAB. However, this reason is only speculation. Determining the true cause will require future research.
Generally, the lactobacilli play a more important role in fermentation processes and effectively promoted lactic acid fermentation for a longer time than lactic acid-producing cocci, eg. enterococci, streptococci, leuconostocs, Weissella and pediococci. When the lactobacilli reach a level of 105 cfu/g FM, silage can be well preserved (Cai et al., 1999c). In this study, the isolated LAB were all lactobacilli and the amount of lactobacilli reached as high as 107 to 108 cfu/g FM. This indicated that TP can be well preserved by ensilaging.
No LAB isolate grew at 5°C. TCP001, TCP004, TCP008, TCP015, TCP017, TCP050, and TCP024 grew well at 10°C. The results will be valuable for the future design of appropriate inoculants for silage fermentation in cold areas. TCP071 and TCP102 grew at 45°C and 50°C. TCP029, TCP045, TCP080, and TCP063 grew well at 45°C. These strains were thus more heat tolerant and could be added to silage at high temperature. TCP037 grew weakly at 45°C and failed to grow at 50°C.
All isolates, expect TCP015 and TCP029, grew well in 3.0% NaCl and only TCP008 grew well in 6.5% NaCl. All isolates, except TCP016, TCP029, and TCP037, grew well from pH 3.0 to 8.0, indicating that most isolated strains were resistant to acid and could grow well during the production of silage.
All isolates, except TCP016, TCP024, and TCP102, were homofermentative rods that did not produce gas from glucose. In contrast to lactic acid-producing cocci, lactobacilli (Rodhe, 1990) are important promoters of lactic acid fermentation for longer fermentation periods. Many studies (Cai et al., 1998; 1999b) have reported that the inoculation of forage with homofermentative lactobacilli, such as Lactobacillus casei and Lactobacillus plantarum, promotes lactic acid fermentation and improves silage quality. However, the heterofermentative weissellas and leuconostocs do not improve fermentation and might cause fermentation loss (Cai et al., 1998).
The species and characteristics of epiphytic LAB can change and influence fermentation losses and silage quality (Li et al., 2011), but the populations of epiphytic LAB are not always sufficiently large or of suitable composition to promote efficient fermentation under farm conditions (Fenlon et al., 1995; McAllister et al., 2002). Thus, our studies aimed at obtaining high-quality silage through the development of additives that stimulate and direct the fermentation process.
In addition, many isolated strains in this study have been demonstrated to be good inoculators of forage. For example, Zheng et al. (2011b) reported that Lactobacillus fermentum produced a large amount of lactic acid to reduce the pH rapidly to below 4.5 which would inhibit Clostridia and enterobacteria. Winters et al. (2000) examined the effect of inoculation with Lactobacillus plantarum and Lactobacillus paracasei subsp. paracasei during ensilage of sterile and nonsterile ryegrass and found that they had a significant effect on silage amino acid profiles. Tabacco et al. (2011) noted that silage inoculated with Lactobacillus buchneri had a lower concentration of lactic acid, lactic-to-acetic acid ratio, and yeast count and higher aerobic stability compared with untreated silage. Inoculation with Lactobacillus buchneri reduced the yeast count to <2 log cfu/g of silage in 16 of 21 farm silage samples, confirming the ability of this inoculum to enhance the aerobic stability of corn silage in farm bunker silos.
Summarily, therefore, some isolates from this study (including Lactobacillus fermentum, Lactobacillus plantarum, Lactobacillus paracasei subsp. paracasei and Lactobacillus buchneri) have already been used as good silage inoculation in the past, producing significant different effects in improving fermentation and preservation. A comprehensive knowledge of the biochemical and physiological properties of both these low and high temperature-tolerant and other isolates should hopefully provide a platform for the discovery and design of inoculants aimed at improving silage fermentation and quality under various prevailing conditions.
16S rRNA gene sequence analysis
A phylogenetic tree of the isolated strains was constructed from evolutionary distances using the neighbor-joining method, as shown in Figure 1. All strains were clustered in the genus Lactobacillus (Figure 1).
Based on phylogenetic analysis, strains TCP102, TCP029, TCP017, TCP001, TCP073, TCP024, TCP037, TCP016, TCP045, TCP063, TCP080, TCP007, and TCP071 were unambiguously identified as Lactobacillus pantheris, Lactobacillus camelliae, Lactobacillus manihotivorans, Lactobacillus harbinensis, Lactobacillus hilgardii, Lactobacillus parafarraginis, Lactobacillus rapi, Lactobacillus buchneri, Lactobacillus pontis, Lactobacillus vaginalis, Lactobacillus panis, Lactobacillus fermentum and Lactobacillus amylovorus, respectively (bootstrap between 83%–100%).
TCP015 was clustered with Lactobacillus coryniformis subsp. torquens and Lactobacillus coryniformis, with a bootstrap value of 100% and a similarity of 16S rDNA sequence of 99.52% with Lactobacillus coryniformis subsp. torquens and 98% with Lactobacillus coryniformis; thus, TCP015 was identified as Lactobacillus coryniformis subsp. torquens.
TCP004 formed a well-defined cluster with 3 strains (Lactobacillus paracasei subsp. tolerans, Lactobacillus paracasei subsp. paracasei and Lactobacillus casei), and 97% bootstrap values confirmed monophyly. Further, TCP004 appeared to be more similar to Lactobacillus casei and Lactobacillus paracasei subsp. paracasei than Lactobacillus paracasei subsp. tolerans, based on its 16S rDNA sequence, which shared 99.59% similarity with those of Lactobacillus casei and Lactobacillus paracasei subsp. paracasei. Based on carbohydrate fermentation patterns, the pattern of strain TCP004 was an exact match to that of the Lactobacillus paracasei subsp. paracasei-type strain and differed from that of the Lactobacillus casei-type strain. Unlike Lactobacillus paracasei subsp. paracasei, Lactobacillus casei grew with glycerol as a carbon source (Table 3). All other isolates that were the same as TCP004 were identified as Lactobacillus paracasei subsp. paracasei.
TCP008 was placed on the phylogenetic tree together with Lactobacillus plantarum subsp. argentoratensis, Lactobacillus pentosus and Lactobacillus planterum in a 96% bootstrap cluster. TCP008 appeared to be more similar to Lactobacillus plantarum and Lactobacillus pentosus than Lactobacillus plantarum subsp. argentoratensis, based on its 16S rDNA sequence (99.38% similarity). According to Curk et al. (1996), Ennahar et al. (2003) and Pang et al. (2011), members of the Lactobacillus plantarum group, including Lactobacillus pentosus, Lactobacillus plantarum, and Lactobacillus paraplantarum, have similar 16S rRNA gene sequences that differ by only 2 bp. Lactobacillus plantarum and Lactobacillus pentosus belong to the same 16S rRNA phylogenetic group and can only be distinguished by phylogenetic analysis of the 16S–23S large space region or partial sequences of the recA gene (Torriani et al., 2001). In this study, we further defined their carbohydrate fermentation patterns and demonstrated unambiguously that the pattern of strain TCP008 was an exact match of that of the Lactobacillus plantarum-type strain and differed from that of the Lactobacillus pentosus-type strain. It appeared that although the Lactobacillus plantarum-type strain produced acid from melezitose, d-raffinose, and α-methyl-d-mannoside, the L. pentosus-type strain did not. Conversely, unlike Lactobacillus plantarum, Lactobacillus pentosus grew with glycerol or d-xylose as a carbon source (Table 3). All other isolates that were the same as TCP008 were identified as Lactobacillus plantarum.
TCP050 appeared to be linked equally to Lactobacillus helveticus and Lactobacillus gallinarum (98% bootstrap), and its 16S rDNA sequence shared 99.23% similarity with Lactobacillus helveticus and 99.37% similarity with Lactobacillus gallinarum. But, based on carbohydrate fermentation patterns, the pattern of strain TCP050 was an exact match to that of the Lactobacillus helveticus-type strain and differed from that of the Lactobacillus gallinarum-type strain.
This study has demonstrated that TP could be well preserved by ensilaging as evident in the virtual maintenance of its original color and the minimal content of unwanted microbes (such as Clostridia and molds) during and at the end of the storage. This coupled with the chemical composition and fermentation characteristics of TPS could make TPS an economic highly nutritious forage alternative. Furthermore, to the best of our knowledge, this is the first isolation of Lactobacillus harbinensis, Lactobacillus manihotivorans, Lactobacillus helveticus, Lactobacillus camelliae, Lactobacillus pontis, Lactobacillus amylovorus, Lactobacillus hilgardii, Lactobacillus pantheris, Lactobacillus panis, Lactobacillus vaginalis and Lactobacillus rapi from silage. Both these and the remaining isolates could form a platform for future design of appropriate inoculants for silage fermentation.
ACKNOWLEDGMENTS
The work was supported by the China Agriculture Research System (CARS-37).


