 |
 |
| Anim Biosci > Volume 37(3); 2024 > Article |
|
Abstract
Objective
The objective of this study was to investigate the regulation relationship of Ten-eleven translocation 1 (Tet1) in DNA demethylation and the proliferation of primordial germ cells (PGCs) in chickens.
Methods
siRNA targeting Tet1 was used to transiently knockdown the expression of Tet1 in chicken PGCs, and the genomic DNA methylation status was measured. The proliferation of chicken PGCs was detected by flow cytometry analysis and cell counting kit-8 assay when activation or inhibition of Wnt4/β-catenin signaling pathway. And the level of DNA methylation and hisotne methylation was also tested.
Results
Results revealed that knockdown of Tet1 inhibited the proliferation of chicken PGCs and downregulated the mRNA expression of Cyclin D1 and cyclin-dependent kinase 6 (CDK6), as well as pluripotency-associated genes (Nanog, PouV, and Sox2). Flow cytometry analysis confirmed that the population of PGCs in Tet1 knockdown group displayed a significant decrease in the proportion of S and G2 phase cells, which meant that there were less PGCs entered the mitosis process than that of control. Furthermore, Tet1 knockdown delayed the entrance to G1/S phase and this inhibition was rescued by treated with BIO. Consistent with these findings, Wnt/β-catenin signaling was inactivated in Tet1 knockdown PGCs, leading to aberrant proliferation. Further analysis showed that the methylation of the whole genome increased significantly after Tet1 downregulation, while hydroxymethylation obviously declined. Meanwhile, the level of H3K27me3 was upregulated and H3K9me2 was downregulated in Tet1 knockdown PGCs, which was achieved by regulating Wnt/β-catenin signaling pathway.
In the germ lineage, changes in DNA methylation/demethylation occur during the formation of primordial germ cells (PGCs) to reset the epigenome of future gametes [1,2]. The TET family members (TET1, 2, and 3) have the capacity to oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which exerts important roles in epigenetic reprogramming in early embryos and PGCs [3,4]. In our previous study, knockdown of Ten-eleven translocation 1 (Tet1) showed impaired survival and proliferation in PGCs, as well as increased 5mC level and reduced 5hmC level [5]. However, the detailed mechanism of this process has not yet been well characterized in chicken PGCs. The present study focused on the molecular regulation of Tet1 mediating demethylation in proliferation and pluripotency in chicken PGCs.
DNA methylation is an important epigenetic modification, which plays a vital role in gene regulation and transposable element silencing [6]. How DNA methylation is reversed is a central question in epigenetic reprogramming [7]. TET1 contains a core catalytic domain and a chromatin-associated CXXC domain to bind CpG sequences which play essential roles in PGC development [8]. The expression of Tet1 is upregulated in reprogramming germ cells, and the locus-specific DNA demethylation in genes related to meiosis was affected by targeted deletion of Tet1 in mice [8,9]. Increasing evidence indicates that TET1 is involved in the epigenetic regulation of proliferation and differentiation in various cells such as embryonic stem cells (ESCs), adult neural progenitor cells, muscle progenitor cells, and cancer cells [10,11]. Mouse PGCLCs effectively recapitulated the genome-wide DNA demethylation occurring in the intragonadal PGCs [12].
Wnt/β-catenin signaling pathway plays a crucial role in PGC development, including specification, migration, and proliferation in PGCs [13]. A recent study demonstrated that loss of TET1 induced aberrant DNA methylation and exhibited suppressor function through regulating Wnt signaling pathway in colon cancer cells [14]. JW74, a β-catenin inhibitor, was diminished through altering histone modifications [15]. JW74 stabilizes AXIN, a component of the β-catenin degradation complex, eventually causing β-catenin degradation [16], whereas BIO led to target gene expression by stabilizing β-catenin from the degradation complex [17]. However, to which degree the Wnt/β-catenin signaling pathway is activated by Tet1 and how it affects the development of PGCs remains unclear.
DNA methylation patterns and histone marks are globally remodeled in PGCs when they migrate to the genital ridges [18,19]. Factors involved in deposition of epigenetic marks regulate the properties related to self-renewal and pluripotency in PGCs. Trimethylation of histone H3 on lysine 27 (H3K27me3) by polycomb group proteins is involved in several epigenome-remodeling steps [20]. The interplay between H3K27me/PcG and DNA methylation also play important work during PGC expansion and migration [21]. A previous study showed that PGCs undergo genome demethylation via the 5hmC intermediate before an increase in the level of H3K27me3 [21]. There are few available data on the epigenome focused on DNA methylation in chicken germ line [5,18].
In mouse PGCs, a global gain in H3K27me3, concomitant with a loss of H3K9me2, occurs after specification and before the entry of these migratory cells into the gonads, where epigenetic reprogramming takes place [21]. In chicken PGCs, the H3K27me3 global level was greatly reduced, whereas the H3K9me3 level was elevated [20]. From these studies, a wide range of mechanisms for achieving demethylation have been proposed that may operate in vivo in the germline [22]. However, mechanisms coordinating these processes remain unclear.
To gain insights into the mechanism by which Tet1 contributes to the proliferation of PGCs, the early molecular events that underlie demethylation in PGCs were investigated. The results demonstrated that Tet1 was required for 5hmC accumulation, which play essential roles in the regulation of proliferation and pluripotency in chicken PGCs by mediating the Wnt4/β-catenin signaling pathway. The present study revealed the key function of Tet1 in regulating the demethylation of PGCs genome and provided a theoretical foundation of the regulation of germ cell development by using PGCs culture model.
All procedures were implemented according to the Local Experimental Animal Care Committee and approved by the ethics committee of Nanjing Agricultural University (Nanjing, China; SYXK-2019-00085).
Fertilized Hyline chicken eggs were incubated at 38.5°C and 60% humidity in an egg incubator (BSS160; Grumbach, Mücke, Germany) according to the standard operating protocols. PGCs were isolated from embryonic genital ridges at E4.5 using our standard protocol [23]. The culture medium was α-MEM (1257107; Gibco, Thermo Fisher Scientifc, Waltham, MA, USA) supplemented with 15% knockout serum replacement (KSR, 10828-028; Invitrogen, Carlsbad, CA, USA), 10 ng/mL leukemia inhibitory factor, 10 ng/mL human bFGF (060-04543; Wako Pure Chemicals, Osaka, Japan), and 2% chicken embryo extract. Chicken PGCs were cultured in an incubator maintained at 37°C with an atmosphere of 5% CO2 and 60% to 70% relative humidity. Anti-stage-specific embryonic antigen (SSEA-1) antibody was used to identify and sort PGCs.
To test the role of Wnt4/β-catenin in Tet1-mediated PGC proliferation, specific activator 6-bromoindirubin-3′-oxime (BIO, 0, 0.5, 1.0, or 1.5 μM) and inhibitor JW74 (0, 5, 10, or 15 μM) (Sigma-Aldrich Chemical Company, St.Louis, MO, USA) were used to detect their effects. BIO and JW74 were dissolved in dimethyl sulfoxide (DMSO), and the control received the vehicle only with a final DMSO concentration of <0.1% that had no significant effect on cell proliferation and survival.
For RNA interference assays, Tet1-specific siRNAs were designed according to our previous study [5]. Cells were plated in 12-well plates at 5×10 cells per well and transfected with 50 nM indicated siRNAs plus siRNA mate (GenePharma, Shanghai, China) according to the manufacturer’s protocol. PGCs were sorted with fluorescence-activated cell sorting (FACS) after transfection and replated for 48 h before being harvested and used for characterizations and further measurement.
Total RNA was extracted from each sample with Trizol reagent (Invitrogen, USA). Quality and purity were determined by the Nanodrop-2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and the integrity was detected by 1.5% agarose gel-electrophoresis. RNA (500 ng) was reverse transcribed to cDNA after DNase treatment by using 5× All-In-One RT MasterMix (Applied Biological Materials Inc., Richmond, BC, Canada) according to the manufacturer’s instructions. Quantitative real time polymerase chain reaction (qRT-PCR) was performed by using SYBR Premix Ex TaqTM kit (Takara, Dalian, China) on an ABI7500 Fast thermal cycler (Applied Bio-systems Biosystems, Foster City, CA, USA) by following the manufacturer’s instructions. The final PCR reactions contained 10 μL 2×SYBR Premix Ex Taq II (Tli RNaseH Plus), 0.8 μL of forwards and reverse primer (10 mM), 2 μL first-strand cDNA and 6.4 μL H2O. Cycling parameters were 95°C for 30 s, followed by 40 cycles at 95°C for 15 s and 60°C for 30 s. Primer specificity was determined by melt curve analysis. Both no-reverse-transcription and no-template controls were used to confirm a lack of genomic DNA contamination and primer dimerization. All samples were normalized with the β-actin using the comparative cycle threshold method (2–ΔΔCt). Primer sequences are listed in Table 1. Differences with p<0.05 were considered as significantly different.
Chicken PGCs were grown on gelatinised glass coverslips, washed with phosphate-buffered saline (PBS), and fixed in 4% paraformaldehyde for 20 min at room temperature (RT). Cells were permeabilized with 0.55 Triton X-100 in PBS for 30 min at RT, washed with PBS and saturated with 2% bovine serum albumin (BSA) in PBS for 1 h. Then PGCs were incubated with the primary antibody raised against anti-SSEA-1 antibody (1:1,000; Developmental Studies Hybridoma Bank, Iowa, IA, USA), 5mC and 5hmC (1:1,000; Abcam, Cambridge, UK) overnight at 4°C in the blocking solution. After three washes with 0.1% Tween-20 in PBS, PGCs were incubated in Alexa-IgG-568 antibodies, fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin M (IgM), or goat anti-mouse IgG (1:1,000; KPL Inc., Gaithersburg, MD, USA) as the secondary antibody in the blocking solution for 1 h. The nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI, 300 nm; Sigma Aldrich, St. Louis, MO, USA). After a brief wash in PBS, samples were mounted with SlowFade Gold antifade reagent (Invitrogen, USA). For the detection of 5mC and 5hmC, nuclear DNA was denatured with 2 N HCl for 30 min and then neutralized with 100 mM Tris-HCl (pH 8) for 10 min. Finally, the images were visualized using laser-scanning confocal microscopy (Zeiss LSM 700; Carl Zeiss AG, Oberkochen, Germany).
PGCs were sorted by FACS after treatment and lysed in radioimmunoprecipitation assay lysis buffer supplemented with protease inhibitors (Complete; Roche, Shanghai, China) and 0.2 nM phenylmethyl sulfonyl fluoride. Protein (20 ng) was denatured and subjected to 10% sodium dodecyl sulfate/polyacrylamide gel electrophoresis. After transfer to polyvinylidene fluoride membrane, the membrane was incubated in 5% dry skim milk in tris buffered saline (0.1% Tween-20) at RT for 1 h. The primary antibodies were rabbit anti-phospho β-catenin (1:1,000; Abcam, UK), rabbit anti-WNT4 (1:1,000; Abcam, UK) and β-actin (1:5,000; Novus Biologicals, Littleton, CO, USA) at 4°C overnight. After 1 h incubation with anti-mouse IgG or anti-rabbit IgG peroxidase-conjugated second antibody (1:5,000; Santa Cruz, Santa Cruz, CA, USA), blots were developed by enhanced chemiluminescence (Thermo Fisher Scientific, Waltham, MA, USA) and exposed to a ChemiDoc-XRS System (Bio-Rad, Hercules, CA, USA) to detect chemiluminescence.
The proliferation of PGCs was determined using the cell counting kit-8 (CCK8) assay (Dojindo, Kyushu, Japan) according to the manufacturer’s protocol. Briefly, 1×104 cells were seeded into 96-well plate and transfected with siRNAs. After transfection for 48 h, fresh culture medium with 10 mL CCK8 solution was added and incubated for another 2 h at 37°C. The optical density (OD) of each well was measured at 450 nm using an automated microplate reader (Thermo Scientific Multiskan GO microplate spectrophotomete).
For the cell cycle analysis, PGCs were first dissociated into single cells and fixed in 70% ethanol at −20°C for 30 min after washing with ice-cold PBS. The cells were incubated with staining solution (50 μg/mL propidium iodide, 0.1% Triton X-100 and 100 mg/mL RNase A in PBS) for 15 min at 4°C. The stained cells were analyzed by flow cytometry (BD FACSCELLULAR, Franklin Lakes, NJ, USA). Data was analyzed using FACSDiva and Modfit LT cell-cycle analysis software (Verity Software House, Topsham, ME, USA).
To detect 5mC and 5hmC levels, genomic DNA was extracted by phenol-chloroform-isoamyl alcohol, and the concentration of samples was serially diluted twofold. Dot blot analysis was performed as described previously [5]. Subsequently, genomic DNA was spotted on a positively charged nylon membrane (Roth, Karlsruhe, Germany), air dried for 15 min and cross-linked using the UV light (20 s, 1,200 J/cm2). Then membranes were blocked in 5% non-fat milk in PBS/0.1% Tween-20 for 1 h at RT before incubation incubated with anti-5mC (1:5,000; Abcam, UK) antibody or anti-5hmC (1:5,000; Active Motif, Carlsbad, CA, USA) overnight at 4°C followed by washing in PBS for 3 times. The membrane was then incubated with HRP-labeled anti-mouse/rabbit secondary antibody (1:5,000; Santa Cruz, USA) and signal was developed using an enhanced chemiluminescence reagent (Bio-Rad, USA). The intensity of signal was measured using Image J software and calibrated against the linear range of the standard curves to estimate the quantities of 5mC or 5hmC in each sample.
Statistical analysis was performed using SAS version 9.4 software (SAS Institute, Cary, NC, USA). When a significant main effect was detected by analysis of variance using SAS, the least-squares analysis was used to compare the different treatments. Differences between control and treatment groups were deemed to be significant when p<0.05.
To investigate the role of Tet1 in the pluripotency of PGCs, siRNA targeting Tet1 was used to transiently knockdown of Tet1 in chicken PGCs. Both mRNA and protein expression of Tet1 in PGCs was examined after Tet1 siRNA transfection for 48 h. The results demonstrated that the mRNA expression level of Tet1 decreased approximately by 50% in Tet1 knockdown group compared with control (Figure 1A) (p< 0.01). In addition, the reduction of TET1 protein was observed by western blotting analysis (Figure 1B). Introduction of siRNA resulted in a 3-fold reduction in the level of TET1 (Figure 1C) (p<0.01). Furthermore, the expression of Nanog, PouV, and Sox2 was declined significantly after the down-regulation of Tet1 (Figure 1D) (p<0.05).
To address the involvement of Tet1 in the level of genomic DNA methylation, the status of 5mC and 5hmC in PGCs was measured by immunostaining and DNA dot blot. Results showed that knockdown of Tet1 led to a general increase in level of 5mC and the level of 5hmC diminished in PGCs (Figure 2A). A similar distribution of 5mC and 5hmC in PGCs was confirmed by DNA dot blot analysis. Expectedly, it was found that Tet1 knockdown induced significant increase in 5mC level in siTet1 group compared with control (Figure 2B–2C) (p<0.01). By contrast, knockdown of Tet1 induced significant decrease 5hmC level in siTet1 group (Figure 2B–2C) (p<0.01). This suggested that the potent capacity of PGCs to induce demethylation was restricted in Tet1 knockdown group.
In this study, the level of β-catenin was detected to investigate the possible mechanism underlying the activity of Tet1. The culture medium was added with 0.5, 1.0, and 1.5 μM BIO or 5.0, 10, and 15 μM JW74. Result revealed that only a modest increase in the level of β-catenin was observed in 0.5 μM BIO, while 1 μM BIO showed substantial increase in β-catenin level (Figure 3A) (p<0.05). By contrast, the level of β-catenin remarkably reduced at 10 μM and 15 μm JW74 treated PGCs (Figure 3B) (p<0.05). The level of β-catenin was rescued in Tet1 knockdown group with addition of BIO, but it was still downregulated significantly compared with control (Figure 3C) (p<0.05).
To determine whether Tet1 exerted the proliferative effect through Wnt4/β-catenin signaling pathway, siTet1 was added along with BIO in PGC culture and the cell-cycle phase distribution was first measured. Results indicated that a notable decrease in G2/M phase was observed in cells treated with siTet1 and JW74 (Figure 4A) (p<0.05), demonstrating that PGC proliferation was significantly inhibited by JW74. By contrast, cell cycle distribution analysis showed the accumulation of cells in S and G2/M phase and a decrease in the proportion of cells in G1 phase after incubation with BIO (Figure 4A) (p<0.05). Cell counting showed there to be less cell proliferation in BIO and siTet1 than that in control group or group treated with BIO alone (Figure 4A) (p<0.05), indicating the proliferative effect of Tet1 on chicken PGCs was induced via the Wnt4/β-catenin signaling pathway. Moreover, the proliferative capacity of cultured PGCs was examined by using the CCK8 assay. As shown in Figure 4B, the growth rate of the siRNA and JW74 group decreased significantly compared with the control group, while the BIO and siTet1 combined with BIO group increased PGC proliferation significantly (p<0.05). To further elucidate the effect of Tet1 knockdown on PGCs, the expression of cell cycle and pluripotency related genes was examined. Tet1 knockdown blocked the mRNA expression of cyclin D1/CDK6, and consequently delayed the entrance to G1/S phase (p<0.05) (Figure 4C). However, this inhibition was rescued by the addition of BIO. To further investigate the role of Tet1 in the pluripotency of PGCs via Wnt4/β-catenin signaling pathway, the expression of pluripotency related genes Nanog, Sox2, and PouV was evaluated, in siTet1 and JW74 group, all those three genes were significantly downregulated, while they were significantly elevated in BIO and siTet1 combined with BIO group (Figure 4D) (p<0.05). These results, along with the dot blot results shown previously, confirmed that Tet1 induced the accumulation of 5hmC in PGCs to promote their proliferation and knockdown of Tet1 in PGCs led to inhibiting cell growth via Wnt4/signaling pathway.
To investigate whether Tet1 is required for maintaining the hypomethylated state, the abundance of 5mC and 5hmC in Tet1 knockdown PGCs was estimated by using dot blot. Result reflected an obviously trend for increasing 5mC level in Tet1 knockdown PGCs was observed (Figure 5A) (p<0.05). Tet1 knockdown as well as JW74 significantly impaired the accumulation of 5hmC, whereas addition of BIO resulted in a marked increase in 5hmC level (Figure 5B) (p<0.05). After disrupting the level of 5hmC in PGCs by down-regulating Tet1, there was no significant change of 5hmC status in BIO and siTet1 combined BIO group (Figure 5B) (p<0.05). As shown in Figure 5B, treatment with siTet1 as well as JW74 prevented 5hmC accumulation in PGCs (p<0.05).
Next, whether histone demethylation plays any roles in chicken PGCs, the protein level of H3K9me2 and H3K27me3 were probed by western blot. Interestingly, the H3K27me3 global level was elevated, whereas the H3K9me2 level was greatly reduced in siTet1 group compared to control group, but BIO reduced H3K27me3 level and elevated H3K9me2 level (Figure 5C) (p<0.05). In addition, siTet1 with BIO elevated both H3K9Mme2 and H3K27me3 (Figure 5C) (p< 0.05). Moreover, we found that the level of H3K27me3 was important for chicken PGC proliferation.
DNA methylation is an epigenetic modification that influences gene expression [24,25]. TET family comprises three members have the capacity to regulate 5mC and 5hmC levels [26]. However, the mechanism by which TET1 is involved in methylation status in PGCs and proliferation program remains unclear. In the present study, knockdown of Tet1 exerted an obvious effect on 5hmC accumulation in PGCs, which probably reflected the diminished capacity of these cells to reprogram. As a result, the proliferation of Tet1 knockdown PGCs was impaired.
TET1 is known to promote DNA demethylation, which influences gene transcription, especially the genes related to proliferation and differentiation in ESCs, muscle progenitor cells and cancer cells [27]. TET1 and TET2 proteins are expressed by pluripotent ESCs [11], regulated by Oct4, and have been implicated in DNA demethylation during PGC development [28,29]. Although Tet1-null female mice were recently shown to have reduced number of germ cells, they fail to properly reactivate meiotic genes [29]. Previous study observed that the loss of Tet1 downregulated a cohort of genes involved in the proliferation of adult neural progenitor cells [27]. The reduction of Tet1 significantly suppressed human dental pulp cells (hDPCs) growth, thus inhibited the capacity of hDPCs to differentiates into odontoblasts-like-cells and generated reparative dentin in response to exogenous stimuli or injury [27]. Recent studies of developing mouse PGCs suggested that the progressive loss of DNA methylation in cells from E8 onward occurs in discrete temporal phases in which different genic and intergenic regions are affected [30]. Several studies reported that the paradoxical phenomenon of TET1 causing both the upregulation of differentiation-related genes and downregulation of pluripotency-related genes [1,27]. In addition, many other studies also demonstrated that Tet1 knockdown caused more genes to be upregulated than to be downregulated [9,14].
Specific gene expression mediated by β-catenin activation generally occurs in a cell-context and cell-dependent manner [15,31]. CyclinD1 and Myc are the best-known target genes that enhance proliferation [31]. Furthermore, JW74 treatment caused the downregulation of cell cycle-related genes essential for entry in G1 phase, such as CyclinD1 and CDK6 [31], which is consistent with this study. In addition, depletion of Tet1 had significant effects on PGC proliferation. Recent study demonstrated that Tet1 exerted tumor suppressive effects in CRC cells [14]. The present study further investigated the role of Tet1 in the proliferation of PGCs and suggested that Tet1 affected the proliferation of chicken PGCs via Wnt4/β-catenin signaling pathway.
Despite the apparent effects on gene expression and PGC proliferation, Tet1 depletion also exerted vital effects on DNA methylation. Global DNA demethylation is a distinguishing epigenetic feature of developing PGCs [27]. TET1 plays a crucial role in the DNA demethylation and has a profound impact on cell self-renewal [27]. Numerous studies have proven that TET1 is important in the modulation of PGC formation, embryo development due to it could control active and passive demethylation via different mechanisms [27]. Previous research revealed that Tet1 regulated neural progenitor cell proliferation in adult mouse brain and mice lacking Tet1 exhibited impaired hippocampal neurogenesis accompanied by poor learning and memory [32]. TET-mediated 5mC oxidation counterbalances de novo methylation to ensure their responsiveness to transcriptional activation driven by upstream signaling pathways and in the absence of TET, abnormal methylation led to a dysregulated Lefty-Nodal circuit [33]. In this study, significant change in 5hmC and 5mC levels was detected after Tet1 knockdown. These results determined the loss of Tet1 contributed to aberrant DNA methylation.
To test whether TET dioxygenase activity is required for Wnt signaling pathway and histone modification, knockdown of Tet1 specifically abolished the enzymatic activity in chicken PGCs. A recent study showed that cell-cycle-related and expression-elevated protein in tumor (CREPT) and p15INK4b-related sequence/regulation of nuclear pre-mRNA domain-containing protein 1A (p15RS) regulated cell proliferation and the cell-cycle transition in chicken embryo fibroblast cells by mediating the transcription of Wnt/β-catenin signaling pathway downstream regulatory genes [34]. Long non-coding RNAs (lncRNAs) H19 facilitated proliferation and inhibited apoptosis through modulating Wnt/β-catenin signaling pathway [35]. In this study, it was found that siTet1 inactivated β-catenin in PGCs cultured in vitro. The expression level of H3K9me2 and H3K27me3 changed because of Tet1 knockdown, and a lower level of H3K9me2 and a higher level of H3K27me3 than that of control were observed.
In mice, epigenetic reprogramming of the germ cells specified in the late epiblast occurs when they migrate and settle in the gonads [4,5]. Loss of dimethylation of H3K9 and DNA methylation, concomitant with an enhancement of H3K27 and H3K4 trimethylation and histone acetylation, occurs at E8.5. In avian species, PGCs had a unique chromatin conformation, and the global level of H3K9me3 was higher in PGCs than in chicken ESCs [20]. On the contrary, the level of H3K27me3 was lower in PGCs than in chicken ESCs, and, most importantly, the nuclear distribution of this PTM was strikingly different, with H3K27me3 only detectable at one large spot similar to ESC chromocentres [20]. H3K27me3-based repression may partially replace H3K9me3-based repression, given that differentiation induces a decrease of H3K9me3 global level. PGCs follow a different epigenetic path in which H3K9me3 prevails over H3K27me3.
This study showed that Tet1 played distinct roles in proliferation and pluripotent reprogramming in chicken PGCs. Loss of 5hmC, accumulation of 5mC and reduction of pluripotency-associated genes, as well as drastic decrease of proliferation, were observed with knockdown of Tet1, which indicated that Tet1 was related with the methylation state to regulate the identification and expansion of PGCs in chickens. Collectively, these findings revealed a fundamental mechanism that Tet1 enhanced the proliferation of chicken PGCs via the activation of Wnt4/β-catenin signaling pathway which is achieved through regulating demethylation status.
Notes
Figure 1
Knockdown of Tet1 in PGCs. The mRNA (A) and protein (B)-(C) expression level of Tet1 was measured. (D) The expression of pluripotency-related genes PouV, Nanog, and Sox2 was detected by qRT-PCR. All the results represent the mean±standard deviation of three independent experiments (n = 3). Tet1, translocation 1; PGCs, pimordial germ cells; qRT-PCR, quantitative real time polymerase chain reaction; PouV, POU domain class 5; Nanong, Nanog homeobox; Sox2, Sex-determining region Y-box2. * p<0.05; ** p<0.01.

Figure 2
Effects of Tet1 knockdown on DNA methylation in PGCs. (A) Tet1 knockdown led to the increase of 5mC (green) and reduction of 5hmC (red) levels in PGCs by immunofluorescence. Scale bar, 20 μm. (B)-(C) The level of 5mC and 5hmC in siTet1 PGCs was confirmed by DNA dot blot. All the results represent the mean±standard deviation of three independent experiments (n = 3). Tet1, translocation 1; PGCs, primordial germ cells; 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine. ** p<0.01.
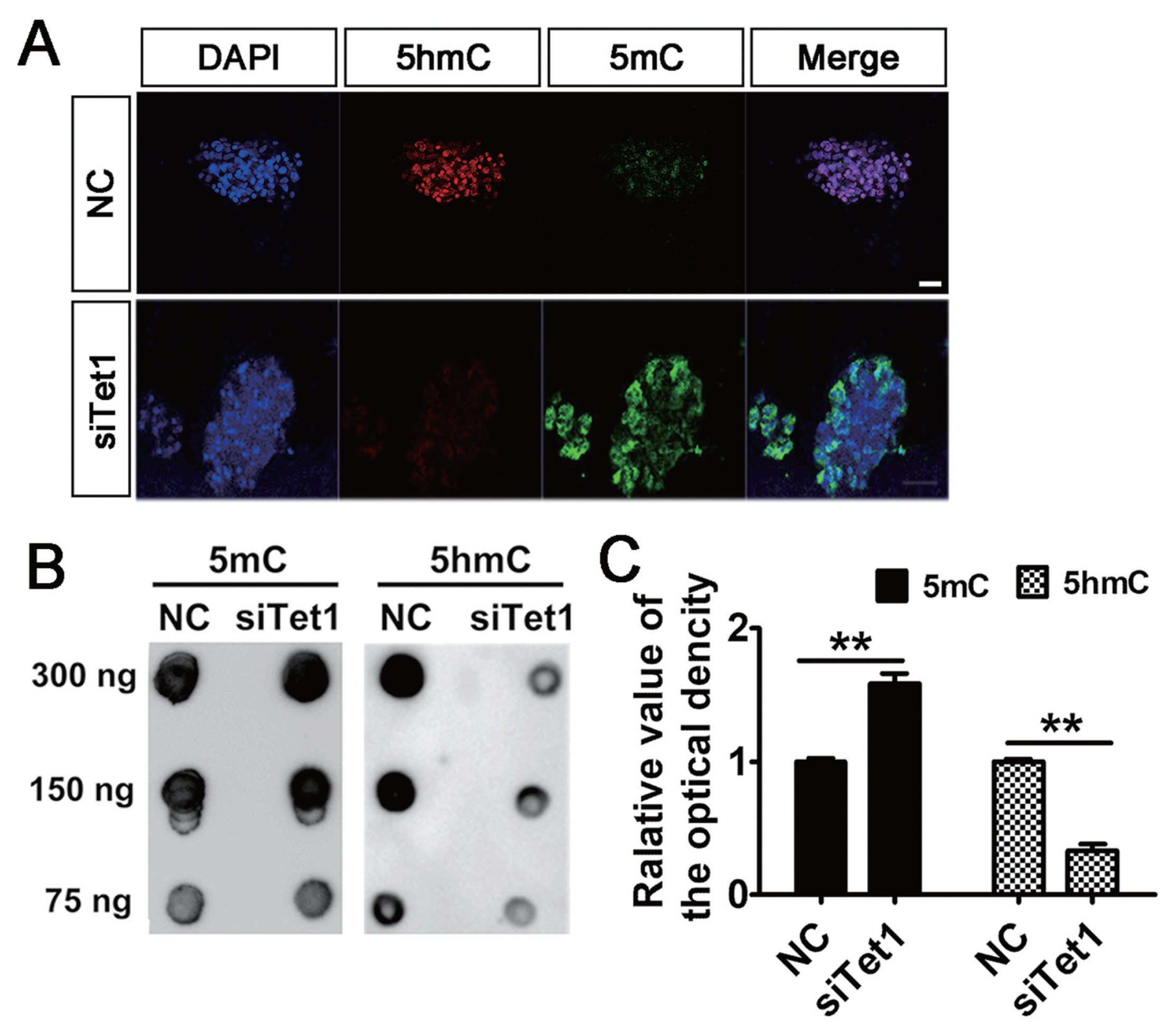
Figure 3
Effects of Tet1 knockdown on Wnt4 and β-catenin protein level. (A)-(B) The level of active β-catenin after BIO or JW74 treatment was determined by western blotting. (C) The protein level of WNT4 and β-catenin in PGCs treated with inhibitors or siTet1 was measured. Tet1, translocation 1; PGCs, primordial germ cells. a–d Means having different letters are different (p<0.05) (n = 3).
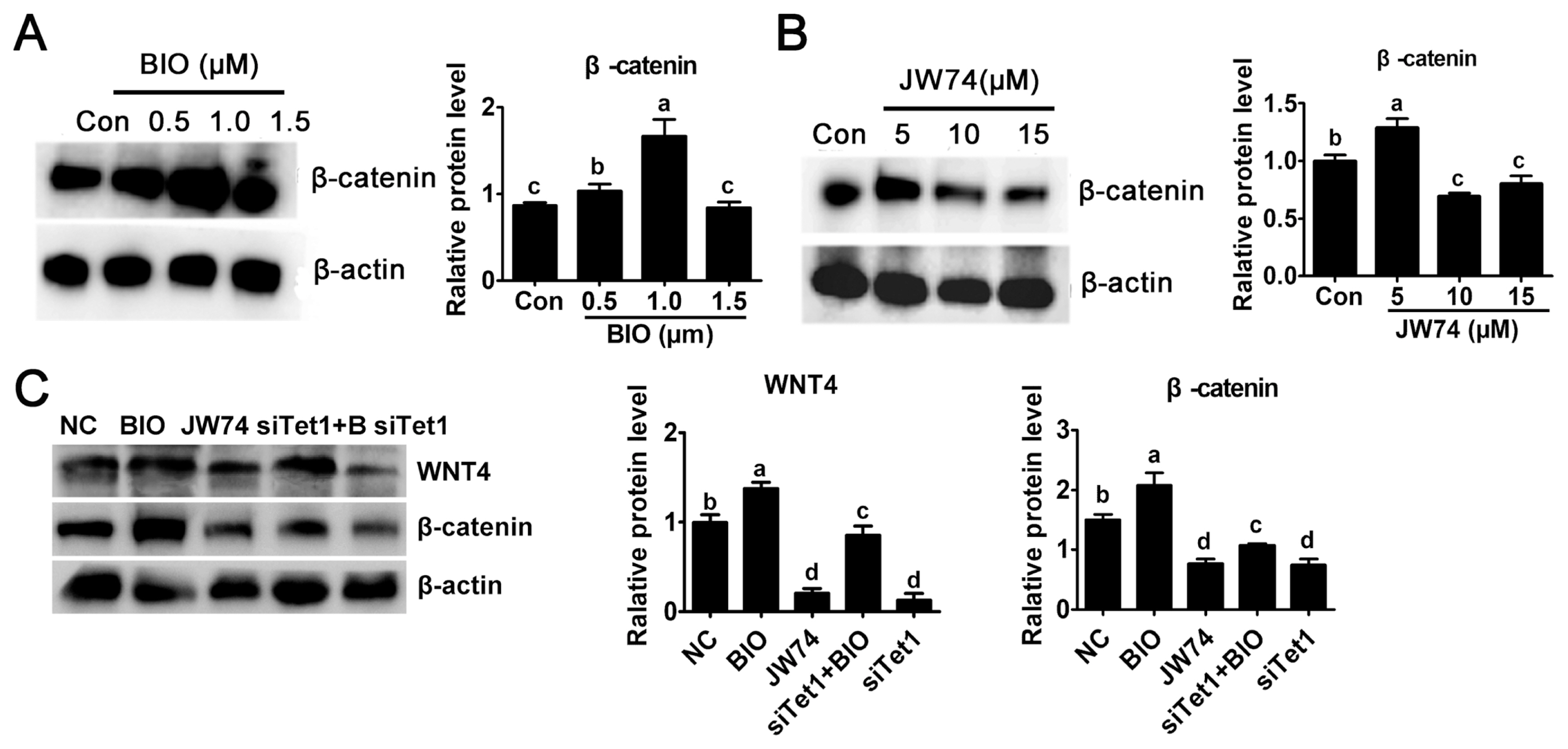
Figure 4
Effects of Tet1 knockdown on PGC proliferation via Wnt4/β-catenin signaling pathway. (A) Detection of cell cycle was conducted in Tet1 knockdown and inhibitor treated PGCs by flow cytometry. (B) Cell growth of each group was measured using the CCK8 assay. (C) The expression of cell cycle-related genes (Cyclin D1 and CDK6) was tested by qRT-PCR. (D) The expression of pluripotency-related genes PouV, Nanog, and Sox2 was detected by qRT-PCR. Tet1, translocation 1; PGCs, pimordial germ cells; CCK8, cell counting kit-8; CDK6, Cyclin-dependent kinase 6; PouV, POU domain class 5; Nanong, Nanog homeobox; Sox2, Sex-determining region Y-box2; qRT-PCR, quantitative real time polymerase chain reaction. a–d Means having different letters are different (p<0.05) (n = 3).

Figure 5
Effects of Tet1 knockdown on DNA and histone methylation. The levels of 5mC (A) and 5hmC (B) in PGCs treated with inhibitors or siTet1 was measured by DNA dot blot. (C) H3K9me2 and H3K27me3 protein level was assessed using western blotting and densitometric evaluation. β-actin was used as an internal control. All the results represent the mean±standard deviation of three independent experiments (n = 3). Tet1, translocation 1; PGCs, primordial germ cells. a–d Means having different letters are different (p<0.05).
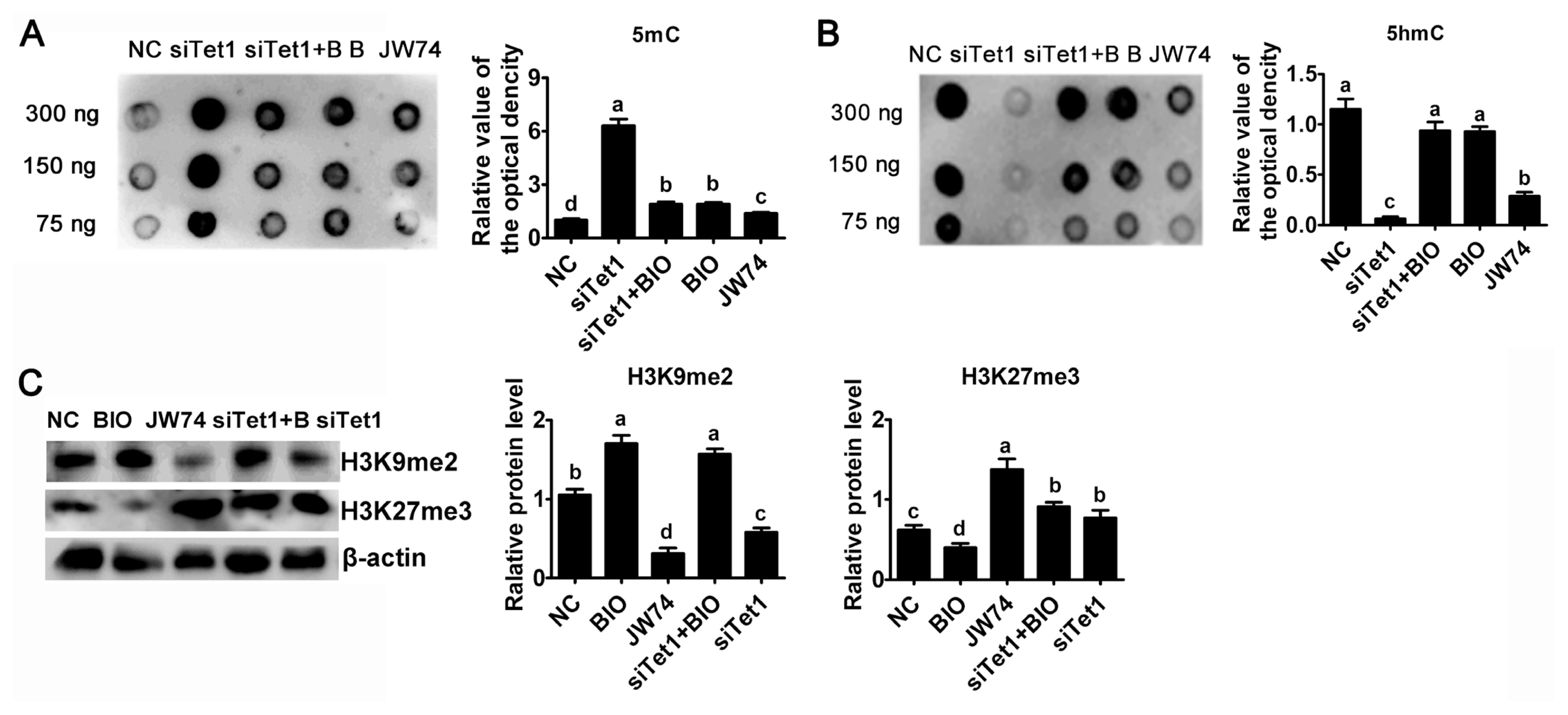
Table 1
The primer sequences for quantitative real time polymerase chain reaction
REFERENCES
1. Ramakrishna NB, Murison K, Miska EA, Leitch HG. Epigenetic regulation during primordial germ cell development and differentiation. Sex Dev 2021; 15:411–31.
https://doi.org/10.1159/000520412


2. Hancock GV, Wamaitha SE, Peretz L, Clark AT. Mammalian primordial germ cell specification. Development 2021; 148:dev189217
https://doi.org/10.1242/dev.189217



3. Wang XX, Ma X, Wei GB, et al. The role of DNA methylation reprogramming during sex determination and transition in zebrafish. Genomics Proteomics Bioinformatics 2021; 19:48–63.
https://doi.org/10.1016/j.gpb.2020.10.004



4. Wang X, Bhandari RK. The dynamics of DNA methylation during epigenetic reprogramming of primordial germ cells in medaka (Oryzias latipes). Epigenetics 2020; 15:483–98.
https://doi.org/10.1080/15592294.2019.1695341



5. Yu ML, Li DF, Cao WY, Chen XL, Du WX. Effects of ten-eleven translocation 1 (Tet1) on DNA methylation and gene expression in chicken primordial germ cells. Reprod Fertil Dev 2019; 31:509–20.
https://doi.org/10.1071/RD18145


6. Gallego-Bartolome J, Gardiner J, Liu WL, Jacobsen SE. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc Natl Acad Sci USA 2018; 115:E2125–34.
https://doi.org/10.1073/pnas.1716945115



7. Zuo QS, Gong W, Yao ZL, Xia Q, Zhang YN, Li BC. Identification of key events and regulatory networks in the formation process of primordial germ cell based on proteomics. J Cell Physiol 2023; 238:610–30.
https://doi.org/10.1002/jcp.30952


8. Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013; 502:472–9.
https://doi.org/10.1038/nature12750



9. Khoueiry R, Sohni A, Thienpont B, et al. Lineage-specific functions of TET1 in the postimplantation mouse embryo. Nat Genet 2017; 49:1061–72.
https://doi.org/10.1038/ng.3868



10. Gavin DP, Chase KA, Sharma RP. Active DNA demethylation in post-mitotic neurons: a reason for optimism. Neuropharmacology 2013; 75:233–45.
https://doi.org/10.1016/j.neuropharm.2013.07.036



11. Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010; 466:1129–33.
https://doi.org/10.1038/nature09303



12. Miyoshi N, Stel JM, Shioda K, Shioda T. Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells. Proc Natl Acad Sci USA 2016; 113:9545–50.
https://doi.org/10.1073/pnas.1610259113



13. Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell 2009; 137:571–84.
https://doi.org/10.1016/j.cell.2009.03.014


14. Kai M, Niinuma T, Kitajima H, et al. TET1 depletion induces aberrant CpG methylation in colorectal cancer cells. PLoS One 2016; 11:e0168281
https://doi.org/10.1371/journal.pone.0168281



15. Jiang X, Tan J, Li JS, et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 2008; 13:529–41.
https://doi.org/10.1016/j.ccr.2008.04.019



16. Waaler J, Machon O, von Kries JP, et al. Novel synthetic antagonists of canonical Wnt signaling inhibit colorectal cancer cell growth. Cancer Res 2011; 71:197–205.
https://doi.org/10.1158/0008-5472.CAN-10-1282


17. Zhang B, Li N, Zhang H. Knockdown of Homeobox B5 (HOXB5) inhibits cell proliferation, migration, and invasion in non-small cell lung cancer cells through inactivation of the Wnt/beta-catenin pathway. Oncol Res 2018; 26:37–44.
https://doi.org/10.3727/096504017X14900530835262



18. Zhang C, Zuo QS, Gao XM, et al. H3K4me2 promotes the activation of lncCPSET1 by Jun in the chicken PGC formation. Animals (Basel) 2021; 11:1572
https://doi.org/10.3390/ani11061572



19. D’Orazio FM, Balwierz PJ, González AJ, et al. Germ cell differentiation requires Tdrd7-dependent chromatin and transcriptome reprogramming marked by germ plasm relocalization. Dev Cell 2021; 56:641–56.
https://doi.org/10.1016/j.devcel.2021.02.007



20. Kress C, Montillet G, Jean C, Fuet A, Pain B. Chicken embryonic stem cells and primordial germ cells display different heterochromatic histone marks than their mammalian counterparts. Epigenetics Chromatin 2016; 9:5
https://doi.org/10.1186/s13072-016-0056-6



21. Mallol A, Guirola M, Payer B. PRDM14 controls X-chromosomal and global epigenetic reprogramming of H3K27me3 in migrating mouse primordial germ cells. Epigenetics Chromatin 2019; 12:38
https://doi.org/10.1186/s13072-019-0284-7



22. Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 2010; 329:78–82.
https://doi.org/10.1126/science.1187945



23. Yu ML, Ge CT, Zeng WD, Mi YL, Zhang CQ. Retinoic acid promotes proliferation of chicken primordial germ cells via activation of PI3K/Akt-mediated NF-kappaB signalling cascade. Cell Biol Int 2012; 36:705–12.
https://doi.org/10.1042/CBI20110542


24. Yamaguchi S, Shen L, Liu YT, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature 2013; 504:460–4.
https://doi.org/10.1038/nature12805



25. Matsui Y, Mochizuki K. A current view of the epigenome in mouse primordial germ cells. Mol Reprod Dev 2014; 81:160–70.
https://doi.org/10.1002/mrd.22214


26. Tahiliani M, Koh KP, Shen YH, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324:930–5.
https://doi.org/10.1126/science.1170116



27. Lian CG, Xu YF, Ceol C, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012; 150:1135–46.
https://doi.org/10.1016/j.cell.2012.07.033



28. Hackett JA, Dietmann S, Murakami K, Down TA, Leitch HG, Surani MA. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Rep 2013; 1:518–31.
https://doi.org/10.1016/j.stemcr.2013.11.010



29. Piccolo FM, Bagci H, Brown KE, et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol Cell 2013; 49:1176
https://doi.org/10.1016/j.molcel.2013.03.011


30. Guibert S, Forné T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res 2012; 22:633–41.
https://doi.org/10.1101/gr.130997.111



31. Lee HC, Lim S, Han JY. Wnt/β-catenin signaling pathway activation is required for proliferation of chicken primordial germ cells in vitro. Sci Rep 2016; 6:34510
https://doi.org/10.1038/srep34510



32. Zhang RR, Cui QY, Murai K, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell 2013; 13:237–45.
https://doi.org/10.1016/j.stem.2013.05.006



33. Dai HQ, Wang BA, Yang L, et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature 2016; 538:528–32.
https://doi.org/10.1038/nature20095


34. Jin K, Chen H, Zuo QS, et al. CREPT and p15RS regulate cell proliferation and cycling in chicken DF-1 cells through the Wnt/beta-catenin pathway. J Cell Biochem 2018; 119:1083–92.
https://doi.org/10.1002/jcb.26277


35. Zhang L, Cheng HL, Yue YX, Li SZ, Zhang DP, He RL. H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/beta-catenin in ox-LDL-stimulated vascular smooth muscle cells. J Biomed Sci 2018; 25:11
https://doi.org/10.1186/s12929-018-0418-4



- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





